A Simple Trick to Showcase Molecular Motion with the Rock Animation
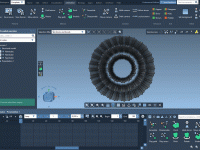
For molecular modelers, visually communicating dynamic behavior is essential—whether it’s for presentations, publications, or team discussions. Yet, it’s often difficult to convey subtle molecular motions effectively and understandably. One recurring challenge? Showing how groups of particles move together in a…