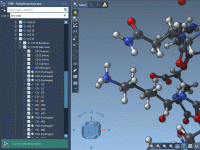
As your molecular models grow bigger and more complex, staying organized becomes essential. Whether you’re aligning protein structures, preparing ligands, or scripting simulations, you’re likely juggling multiple structures, tools, and settings at once. This is especially true when working on…