Author: OneAngstrom
Making Sense of Node Group Selection in SAMSON
Quickly Filter Molecules by Number of Chains in SAMSON
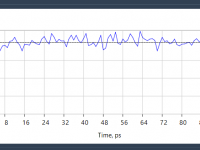
Getting Your Molecular System to the Right Temperature in GROMACS Wizard
Make Atoms Flash: A Simple Way to Emphasize Molecular Events
Manually editing molecules without breaking your simulation? IM-UFF makes it possible.
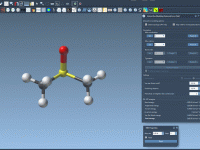
One common challenge in molecular modeling is modifying molecular structures on the fly—connecting or breaking bonds—without having to recalculate everything from scratch. Typically, simulations require a fixed topology, and making any structural change leads to restarting your workflow, setting parameters…