Author: OneAngstrom
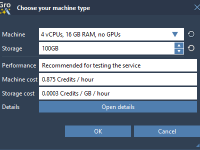
Is Your Local Machine Too Slow? Try Running GROMACS in the Cloud
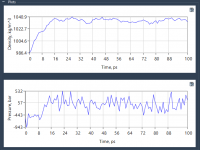
From Chaotic to Stable: Understanding NPT Equilibration in Molecular Simulations
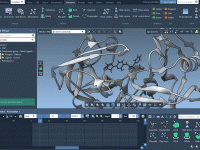
Making Your Molecular Models Disappear—Smoothly
Choosing the Right Unit Cell in GROMACS Simulations
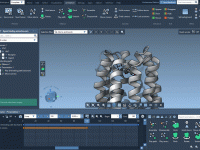
From Motion to Model: Exporting Atom Paths in SAMSON
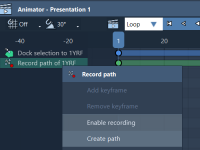
Whether you’re visualizing protein docking, showcasing a molecular simulation, or animating custom atom movements, path recording can drastically enhance molecular presentations. But what happens once the movement is complete? Many molecular modelers face the challenge of capturing and exporting atom…