Author: OneAngstrom
Speed Up Molecular Modeling with Attribute-Based Backbone Filters
When building or analyzing complex molecular systems, molecular modelers often face a recurring challenge: identifying specific fragments such as backbones with particular properties—like charge, composition, visibility, and more—across vast datasets or large molecular assemblies. Manually browsing through molecular groups wastes…
Adding New Molecular Modeling Capabilities to SAMSON with Apps
Managing Note Visibility in SAMSON: How to Hide or Show Annotations Efficiently
A Seamless Way to Add Backgrounds to Molecular Presentations
A Visual Way to Show Molecular Disassembly for Presentations and Learning
A Clearer View of Protein-Ligand Interactions: 2D Meets 3D
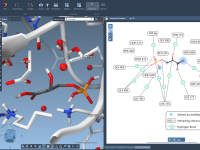
When analyzing protein-ligand complexes, understanding non-covalent interactions—like hydrogen bonds, van der Waals contacts, and pi-stacking—can feel overwhelming in 3D alone. That’s where 2D interaction diagrams make a difference: they simplify complexity and help researchers interpret molecular details more clearly. Interaction…
When Less Is More: Concealing Atoms to Clarify Molecular Animations
How to Easily Customize Metallic, Transparent, and Emissive Materials in Molecular Models
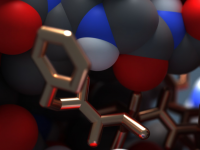
Creating molecular visuals that are both accurate and visually engaging is a common challenge among molecular modelers. While generating molecular structures can be straightforward, rendering them with materials that are scientifically meaningful—or simply aesthetically pleasing—can become time-consuming without the right…