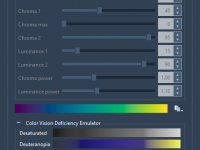
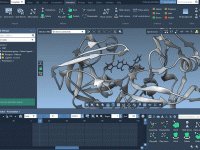
Speeding up Nanotube Design with Pattern Editors in SAMSON
For molecular modelers working on nanostructures, building carbon nanotubes and similar architectures atom by atom is time-consuming and error-prone. Small misalignments between atoms can create structurally incoherent models and increase the time required for cleanup and minimization. If your work…