From Confusing to Clear: Managing Molecular Complexity with Visual Presets
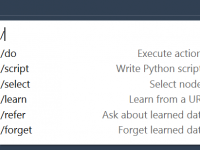
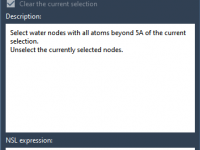
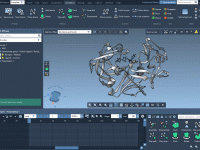
Visualizing molecular systems often feels like a balancing act—switching representations, adjusting colors, highlighting key atoms—before you’ve even started analyzing. For researchers and students dealing with increasingly large biomolecular structures, the time spent configuring these visualizations manually can quickly add up.…