Author: OneAngstrom
Efficiently Controlling Label Visibility in Molecular Models
A Practical Guide to Custom Index Groups in GROMACS Wizard for Better Control and Analysis
Running GROMACS in SAMSON with Your Own Installation: Why and How
How to Find Nearby Atoms Using Proximity in SAMSON
Choosing the Right Visual Model in SAMSON: A Practical Overview
Watching Atoms Move: A Simple Guide to Interactive Simulations in SAMSON
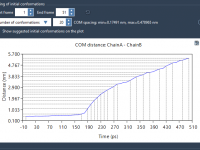
Tuning Protein Conformations with an Interactive Ramachandran Plot: A Visual Approach
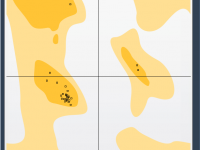
When working with protein structures, even small mistakes in backbone conformation can introduce physical strain or mislead simulation outcomes. Before launching into molecular dynamics or interpreting mechanistic insights, it’s helpful to take a moment and check your model’s backbone—especially when…