Author: OneAngstrom
Make Molecules Fade Away: When ‘Disappear’ Is Better Than Hide
When creating molecular animations for presentations or publications, molecular modelers often face the challenge of showing molecular structures gradually vanish—without making it look abrupt or confusing for the audience. While the classic ‘Hide’ effect can switch off visibility instantly, sometimes…
Filtering Molecular Chains by Attributes in SAMSON’s NSL
Controlling Visibility in Molecular Models: A Hidden Time-Saver
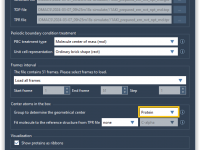
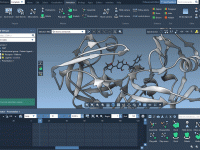
Quickly Align Protein Structures Based on Specific Residue Regions
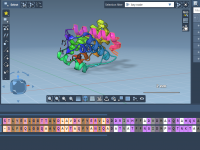
When exploring protein function or designing ligands, it’s often essential to compare specific functional regions, rather than entire protein structures. This is especially true in structure-based drug discovery, homology modeling, and evolutionary studies, where conserved motifs might be hidden among…
Precise Bond Filtering in Molecular Models Using Bond Types
Selecting Cameras with Precision in SAMSON’s Node Specification Language
A Simple Way to Align Molecular Structures in SAMSON
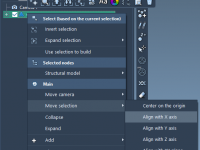
When working on multi-component molecular systems—such as protein-ligand complexes, nanostructures, or assembling ligand libraries—being able to precisely align and position structures is essential. But manual alignment can be both time-consuming and error-prone, especially when scaling operations or ensuring consistency across…