Author: OneAngstrom
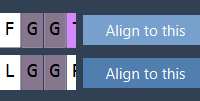
When Whole Protein Alignment Isn’t Enough
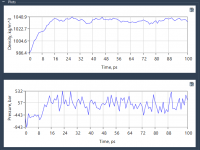
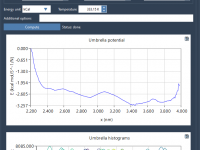
Why Your Density Isn’t Stabilizing: A Practical Guide to NPT Equilibration in SAMSON’s GROMACS Wizard
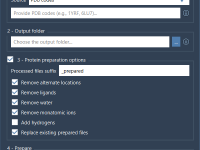
Cleaning Dozens of Protein Structures, Effortlessly
Make Symmetry Work for You: Tips for Visualizing Macromolecular Symmetry
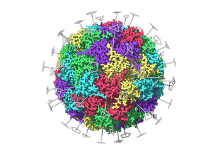
When studying large biomolecular assemblies—such as viruses, protein complexes, or synthetic nanomaterials—symmetry isn’t just aesthetic. Identifying and understanding symmetry can help uncover repeated functional motifs, improve structural validation, and dramatically reduce computational demands. SAMSON’s Symmetry Detection extension brings powerful symmetry…