In drug design and molecular optimization, exploring structural variants of a given parent molecule is a common strategy to improve activity, solubility, or other properties. Manually generating all these variants, however, can be repetitive and error-prone—even before simulations begin.
The Replace fragments feature in the SMILES Manager extension of SAMSON addresses this pain point by offering a simple yet powerful tool to generate libraries of modified molecules through fragment substitution. Whether you’re optimizing a lead compound or performing isosteric replacements, this functionality can help streamline that part of your workflow ⚙️.
What it does
With just a few steps, Replace fragments automatically creates many new molecules by systematically swapping a specific fragment (e.g., an amide group or a ring system) with other specified fragments. This allows rapid exploration of molecular diversity without building new molecules one by one.
How it works
Let’s walk through a typical use case. Suppose you have a molecule like:
|
1 |
COc1cc(CNC(=O)CCCC/C=C/C(C)C)ccc1O |
Your goal: replace the C(=O)NC moiety with a range of alternative fragments for structure–activity relationship studies.
Step 1: Load your initial molecule in the Replace fragments tab.
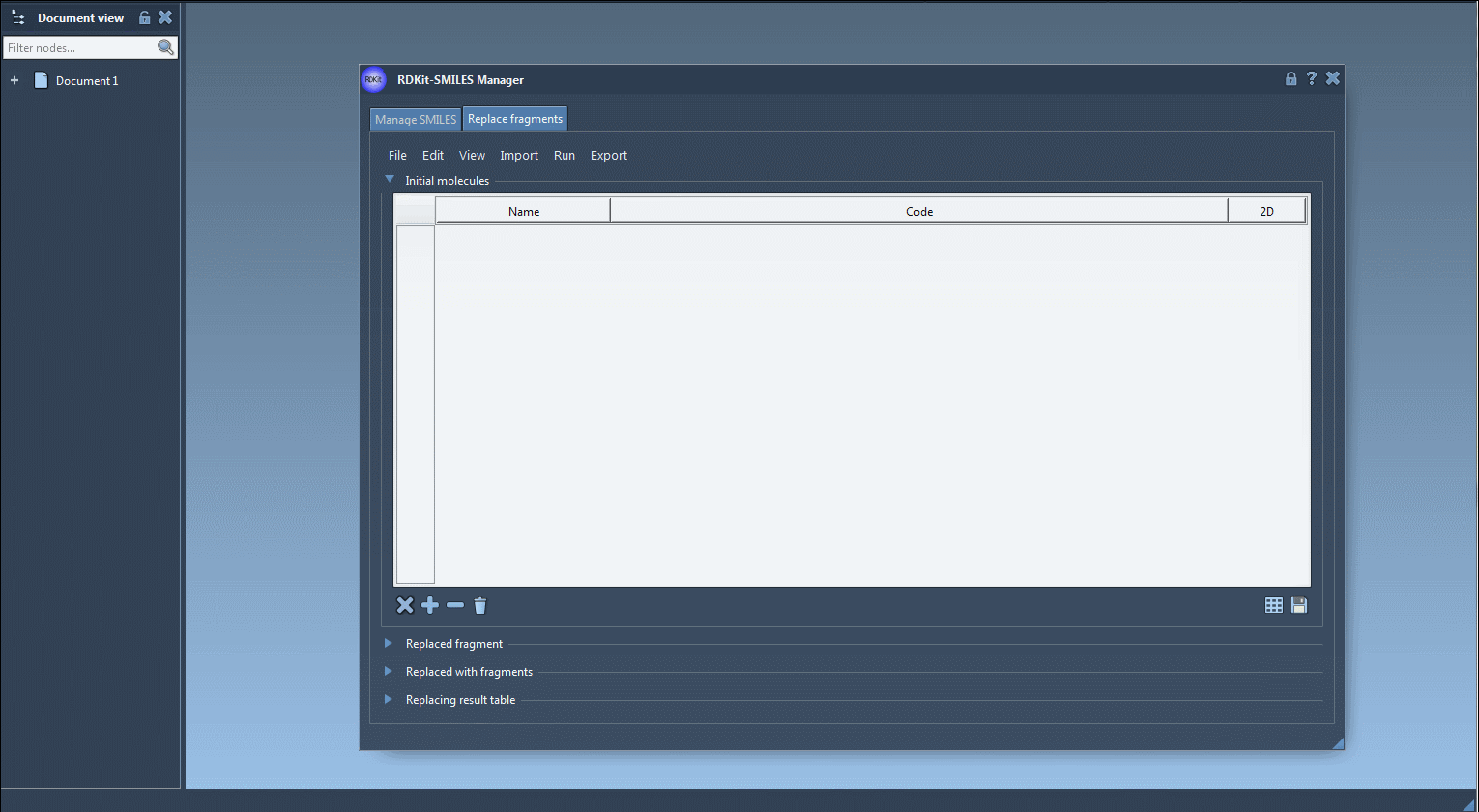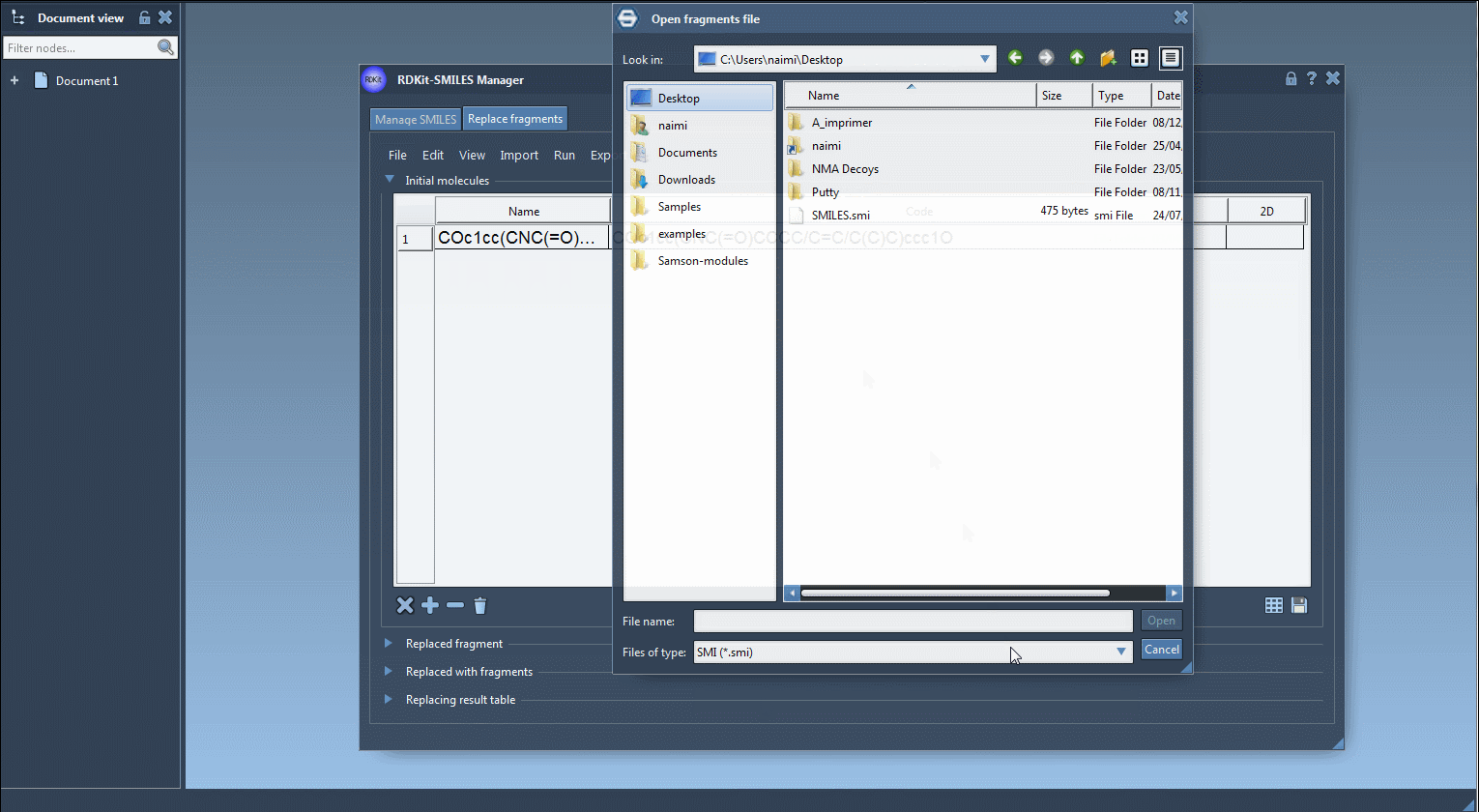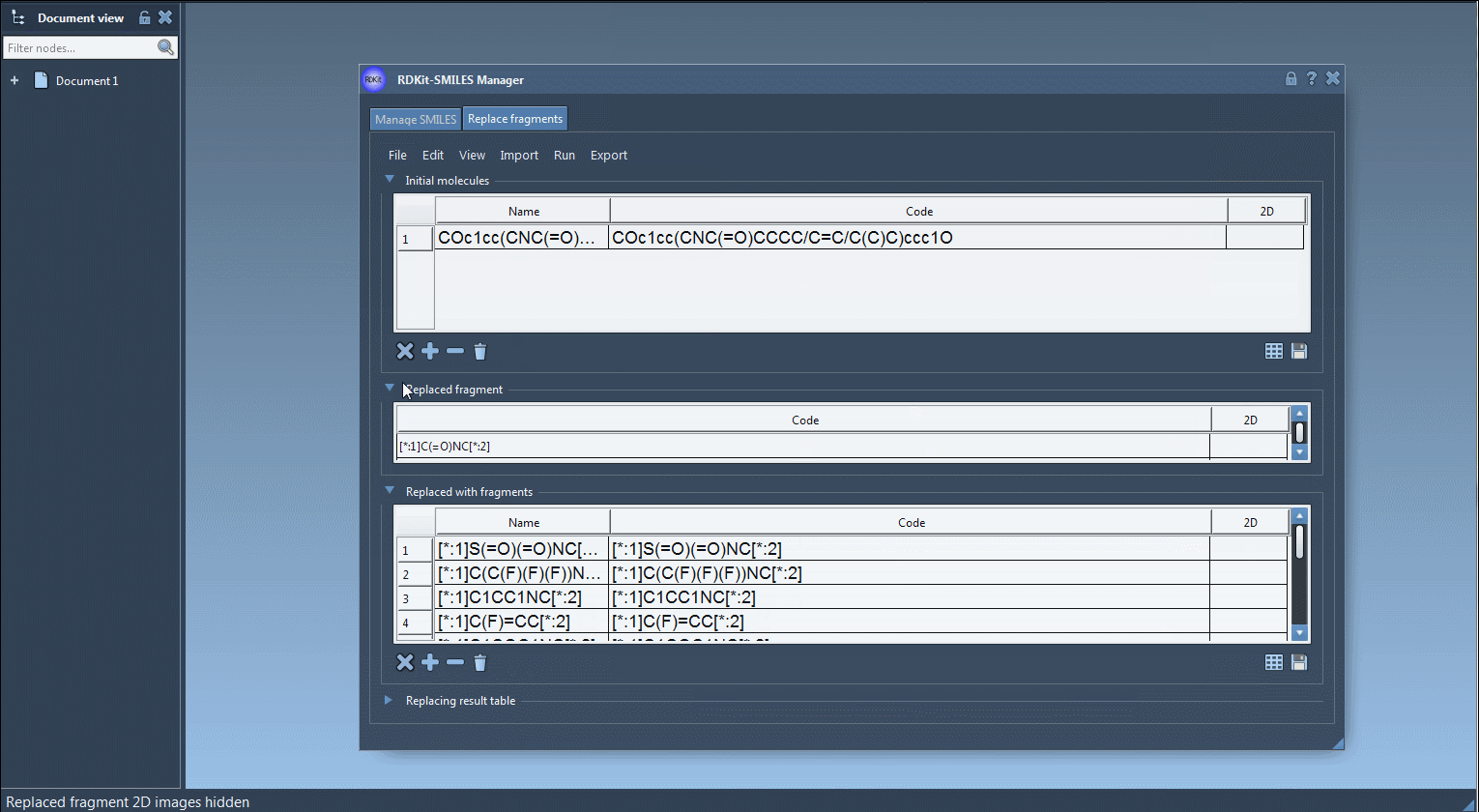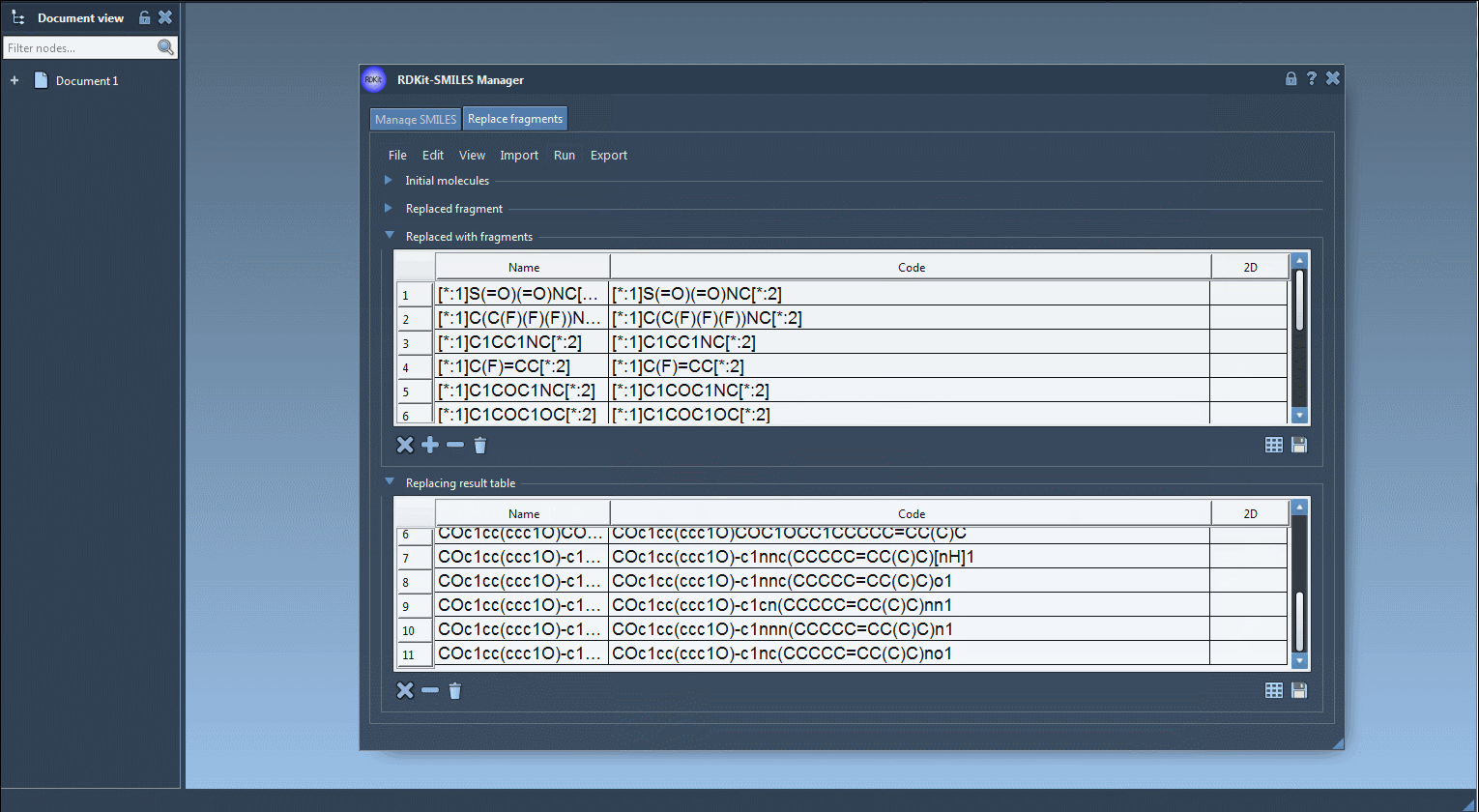
Step 2: Define the fragment to replace. This is done using SMARTS syntax with wildcard anchors:
|
1 |
[*:1]C(=O)NC[*:2] |
Here, [:1] and [:2] connect the fragment to the rest of the molecule, allowing clean integration of the replacement fragments.
Step 3: Provide a list of replacement fragments, for instance:
|
1 2 3 4 |
[:1]S(=O)(=O)NC[:2] [:1]C(C(F)(F)(F))NC[:2] [:1]C1CC1NC[:2] [:1]C(F)=CC[:2] |
You can either enter them manually or load them from a file.
Step 4: Run the replacement process. The generated molecules are displayed in a table, with corresponding names and 2D depictions auto-generated by RDKit.
Step 5: Export the resulting molecules to your SAMSON Document, generate 3D structures, or save them for later analysis as individual images or in a grid.
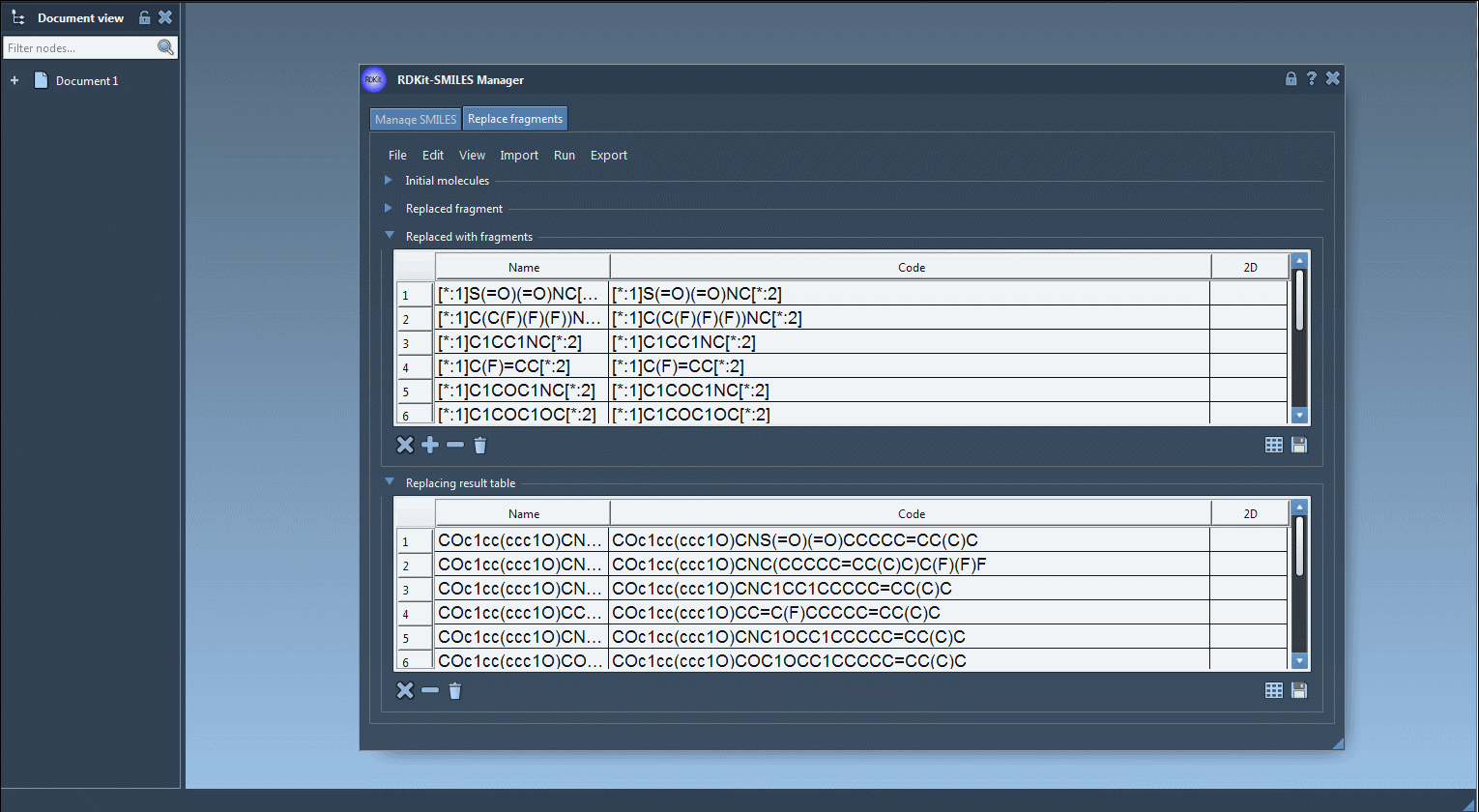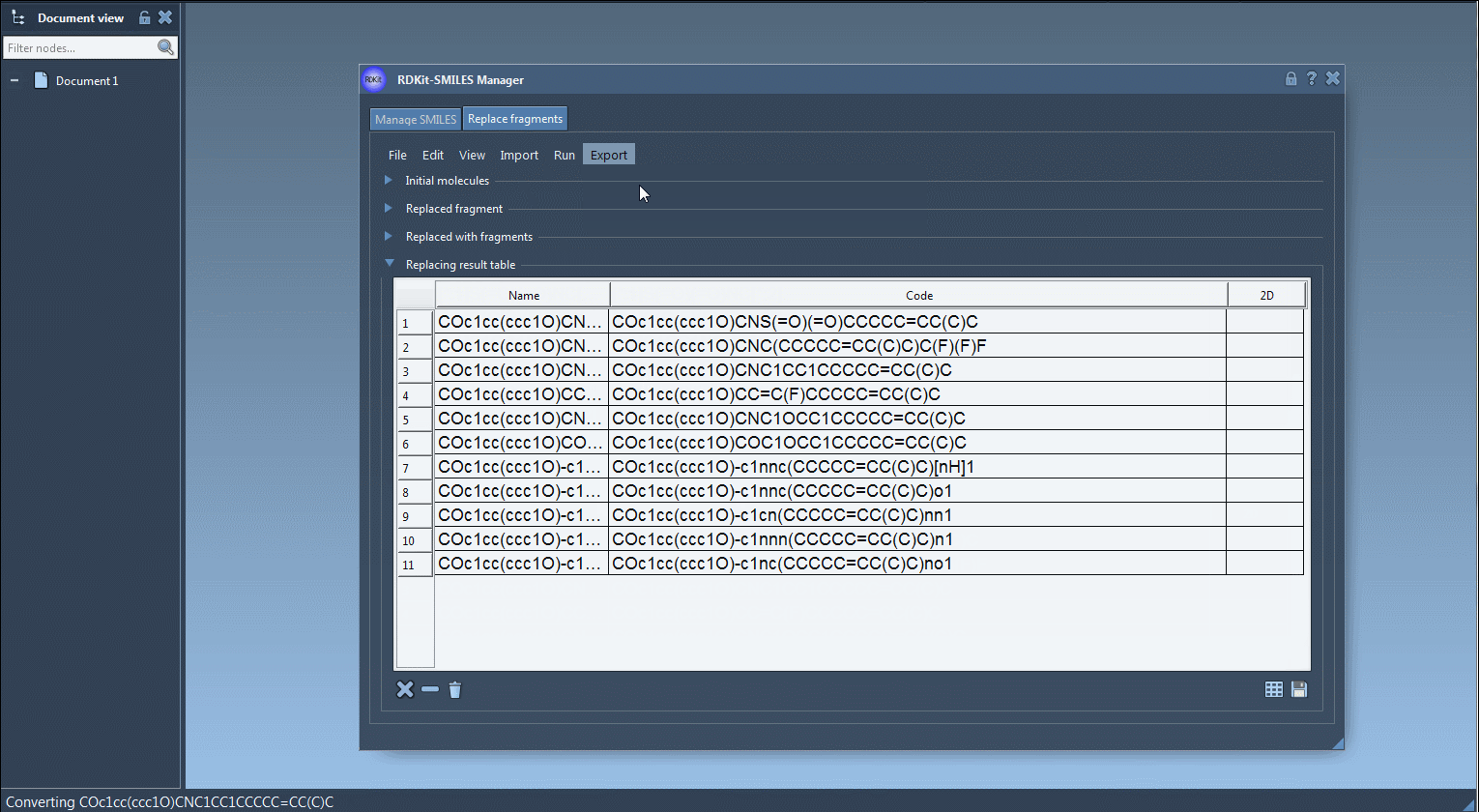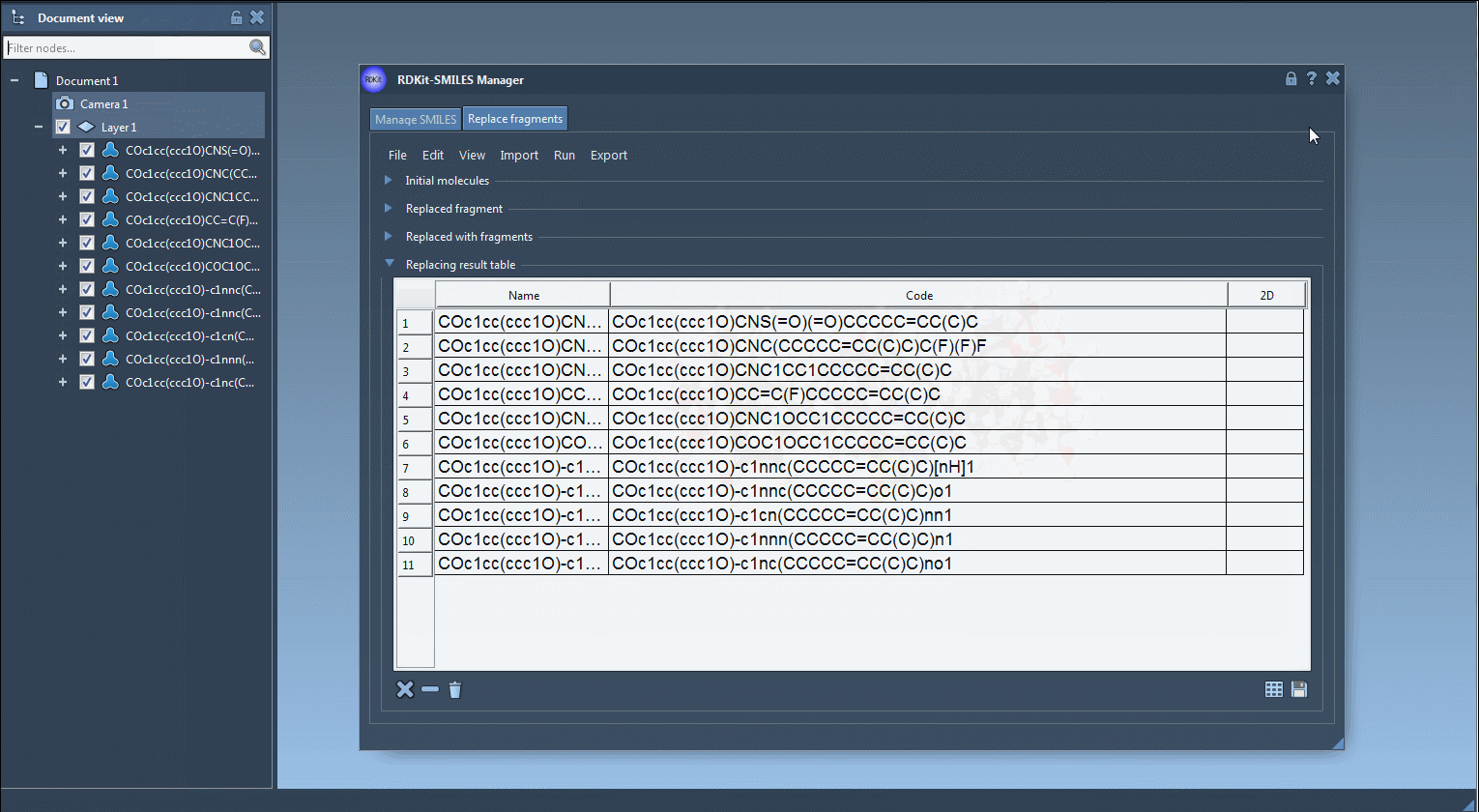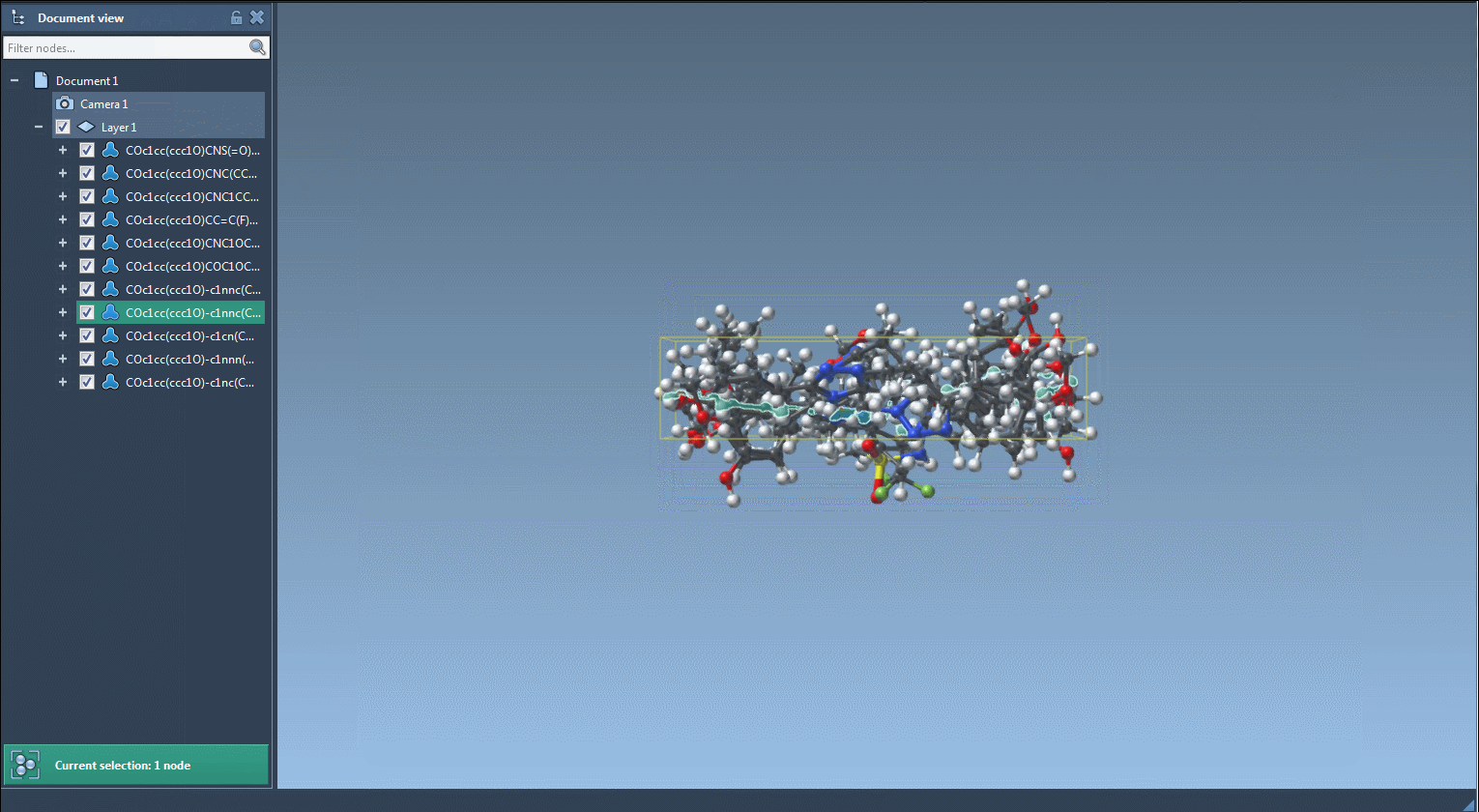
Why it matters
This tool is especially useful when you want to:
- Quickly generate analog libraries for virtual screening
- Explore bioisosteric replacements
- Diversify lead compounds with ease
- Visualize differences using built-in 2D and 3D depiction tools
What’s more, the integration with RDKit means compatibility with widely used cheminformatics workflows, while benefiting from the visual and computational capabilities of SAMSON.
Final thoughts
Whether you’re exploring SAR, identifying better binders, or generating training datasets, the Replace fragments functionality is a valuable addition to your molecule design toolkit. All without writing scripts.
➡️ Learn more in the full documentation.
SAMSON and all SAMSON Extensions are free for non-commercial use. You can get SAMSON at https://www.samson-connect.net.





