Transition pathways are a cornerstone in molecular modeling, as they provide valuable insights into the mechanical and energetic processes between molecular conformations. However, ensuring these pathways are optimized for accuracy and efficiency can be a major pain point for researchers. The Parallel Nudged Elastic Band (P-NEB) method seeks to address this problem by refining minimum energy paths with high efficiency while maintaining equal spacing between intermediate configurations.
Why P-NEB Matters
The Nudged Elastic Band (NEB) method is a powerful approach for identifying saddle points and minimum energy paths between known molecular states. However, optimizing these pathways manually or with non-parallelized methods can be time-consuming and computationally intensive, especially for complex systems. This is where the P-NEB app in SAMSON comes into play. By leveraging parallel execution, the P-NEB method allows for faster and more accurate optimization, even for systems with many configurations.
Step-by-Step: Applying P-NEB to Transition Paths
Here’s how to significantly improve your molecular insights using the P-NEB app:
- Load Your Input Model
Start by loading a trajectory or structure into SAMSON. For example, download a sample document from SAMSON Connect, such as a Zinc ligand unbinding trajectory or a protein-ligand complex of Lactose permease and Thiodigalactosid.

- Start the P-NEB App
In the SAMSON interface, navigate to Home > Apps > All > P-NEB and open the app. Alternatively, use the “Find everything…” search box to locate it quickly.
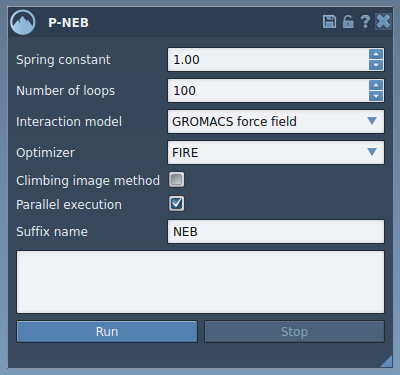
- Configure P-NEB Settings
In the P-NEB interface, fill in the following parameters:
- Spring constant: Use 1.00 for balancing neighboring configurations.
- Number of loops: Set to 100 for optimal energy refinement.
- Interaction model: Select “Universal Force Field.”
- Optimizer: Choose “FIRE” (Fast Inertial Relaxation Engine).
- Climbing image method: Leave unchecked initially.
- Parallel execution: This should always be checked for faster computations.
- Suffix name: Enter “NEB” for easier organization of the resulting path or conformations.
- Run the Optimization
Select a path (or set of conformations) in the Document view, and click the Run button in the P-NEB app. Note: Applying P-NEB to a path is faster than using it on individual conformations. You can convert a set of conformations into a path by selecting them and using the context menu option “Conformation > Create path from conformations.”
Once the computation progresses, you can monitor the optimization via the status bar. Upon completion, a new optimized path (or conformations) will appear in the Document view for further analysis.
Refine and Analyze Your Results
After optimization, you can inspect the new path using the Inspector menu. You can also double-click the path for animations or right-click for context-specific options like “Path > …” in SAMSON. Spend some time analyzing the improved molecular trajectory to extract key insights for your study.
Conclusion
Optimizing molecular transition paths has never been more approachable, thanks to the P-NEB method in SAMSON. With its efficient parallel computation and customizable settings, you can focus on analyzing meaningful results rather than battling computational challenges. To dig deeper into this powerful tool, visit the detailed documentation here.
SAMSON and all SAMSON Extensions are free for non-commercial use. Get SAMSON today at SAMSON Connect.





