One of the common challenges in drug design and lead optimization is efficiently exploring chemical space by generating analogs of a parent molecule. Traditional workflows require scripting or toolchains that are time-consuming and error-prone — not ideal when you’re handling hundreds or thousands of candidate molecules. The SMILES Manager extension in SAMSON offers a practical solution with its Replace fragments feature, making this process more intuitive and visual.
Instead of manually editing SMILES or relying solely on code, you can now quickly generate molecular libraries by defining a fragment to be replaced within an initial molecule, and visually inspecting the substituted derivatives. Here’s how the process works step-by-step, directly within the graphical interface of SMILES Manager:
1. Define Your Starting Point
Begin by importing your initial molecule — either manually or from a document. For instance, you might start with this SMILES string representing a lead compound:
|
1 |
COc1cc(CNC(=O)CCCC/C=C/C(C)C)ccc1O |
2. Choose the Fragment to Replace
Specify the chemical pattern you’d like to replace. Fragments are defined using SMARTS-like syntax with placeholder atoms at the points of attachment. For example:
|
1 |
[*:1]C(=O)NC[*:2] |
This represents an acylated amine segment — a common functional group in bioactive molecules.
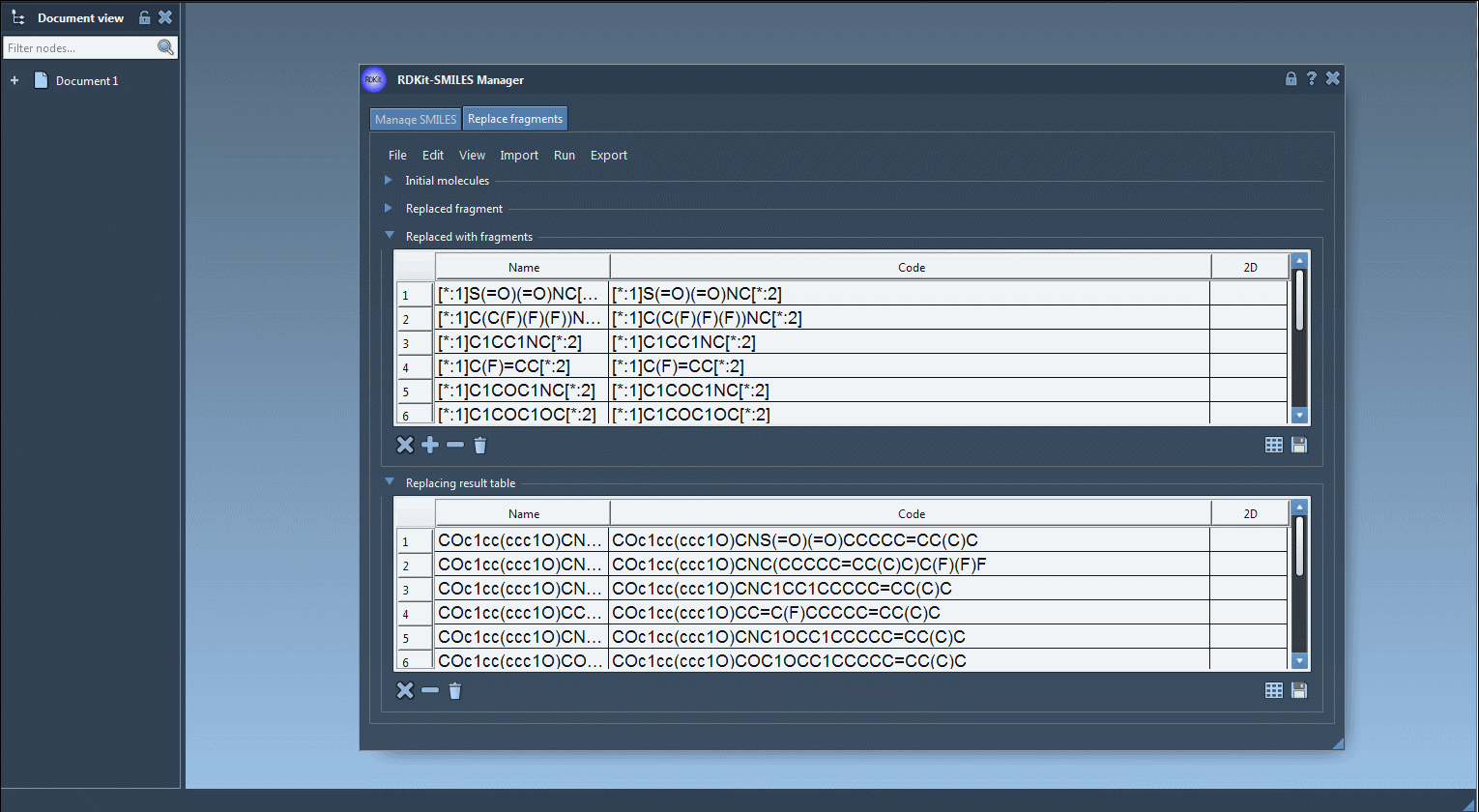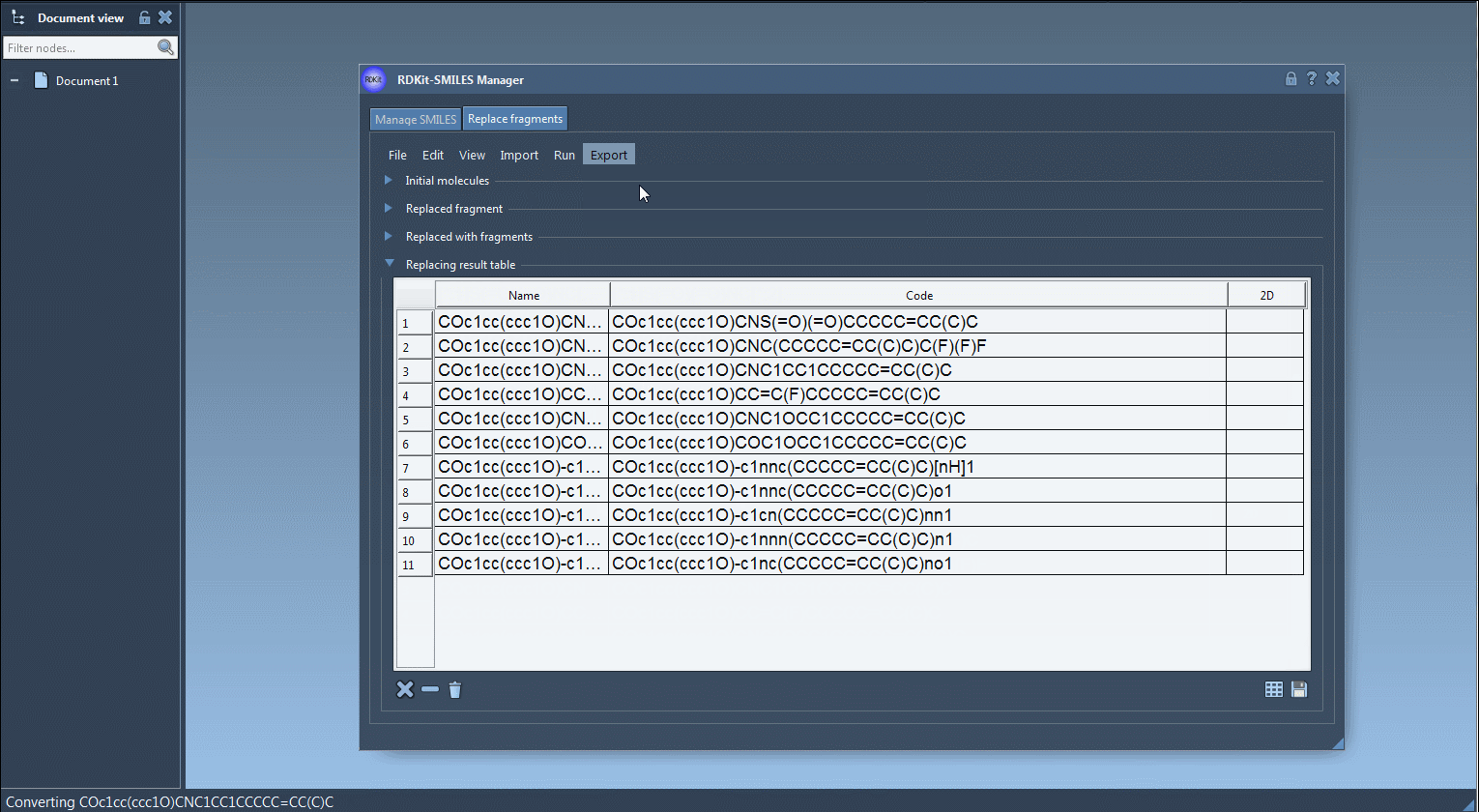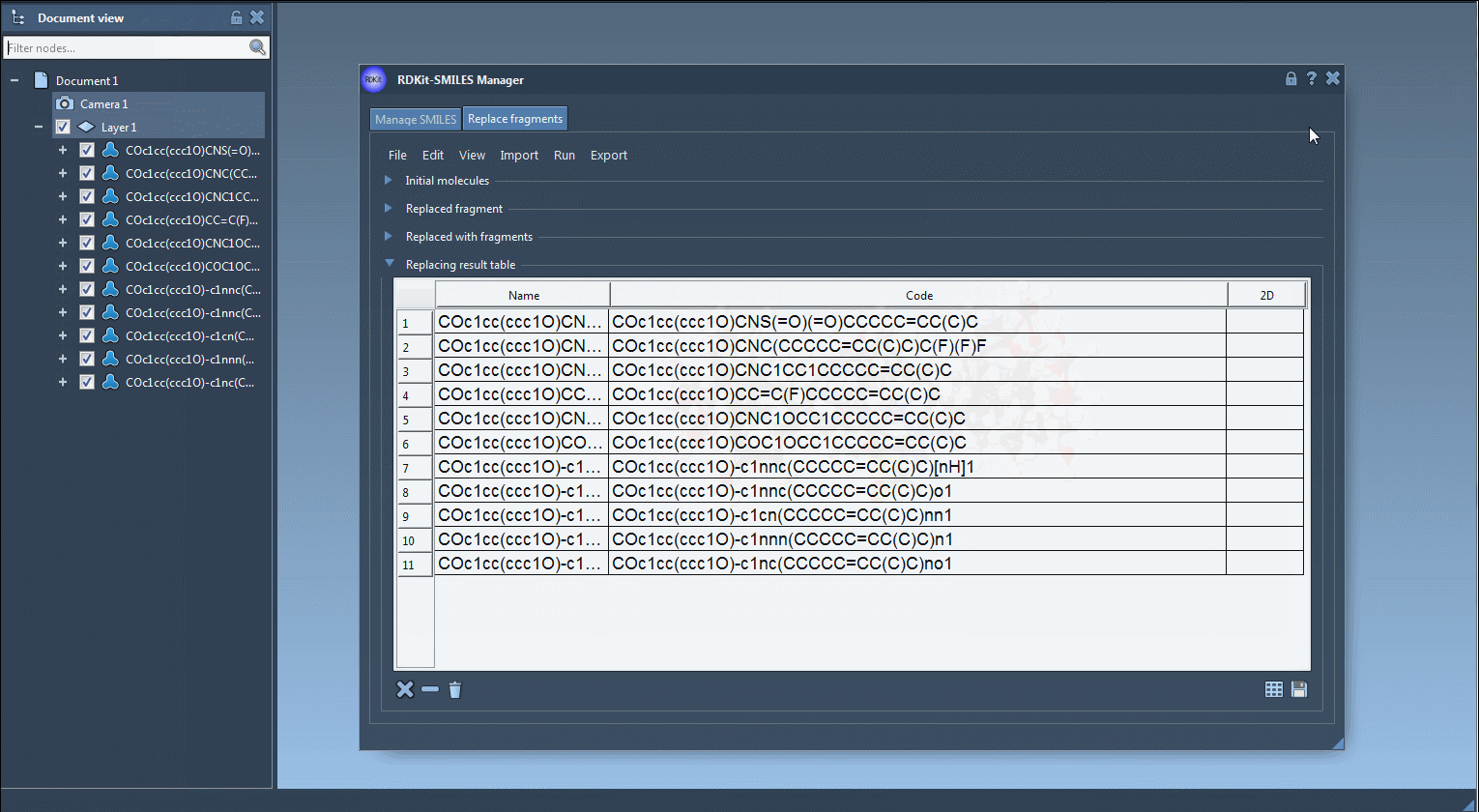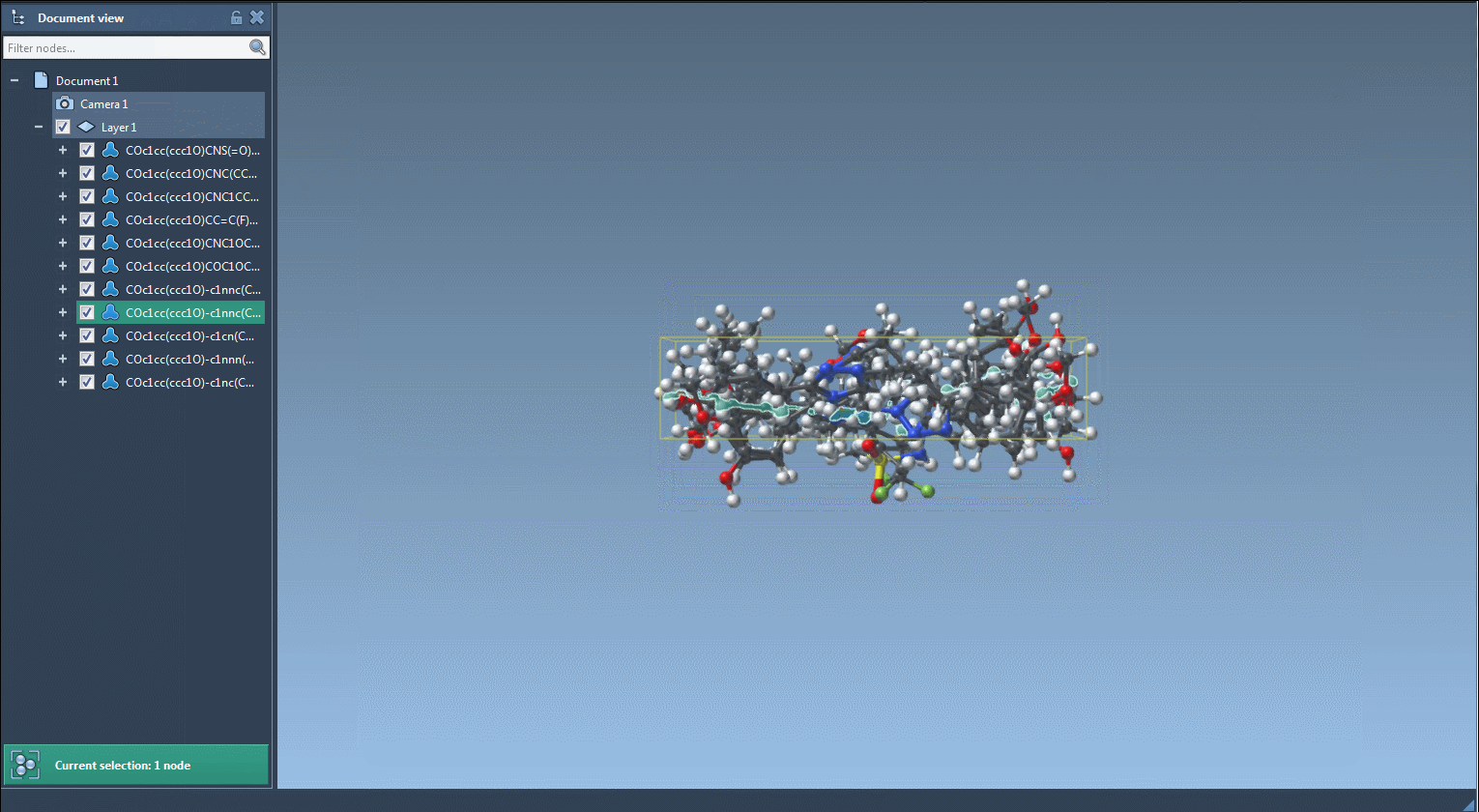
3. Prepare the Replacing Fragments
Enter your replacement fragments in a similar SMARTS pattern, preserving the attachment points:
|
1 2 3 4 5 |
[*:1]S(=O)(=O)NC[*:2] [*:1]C(C(F)(F)(F))NC[*:2] [*:1]C1CC1NC[*:2] [*:1]C(F)=CC[*:2] [*:1]C1COC1NC[*:2] |
You can add these fragments manually or import them from a file. Each replacement will result in a chemically modified analog where the selected fragment is substituted.
4. Generate and Inspect Results
Click on Replace fragments from the Run menu to create your library. The derived molecules are automatically listed, complete with names and generated 2D structures. You can visualize them as a table, zoom into individual depictions, or save them as a grid image:
5. Move to 3D or Export
Once you’re satisfied with your new molecules, you can export them to your SAMSON Document. From there, generate 3D structures, run calculations, or continue designing. You also have options to save the table with 2D depictions or export selected structures as a grid, which is helpful for comparison or presentations.
In Summary
The Replace fragments functionality in SAMSON’s SMILES Manager saves time in molecular design by automating the process of substituent exchange. What once required scripting or batch generation in separate software can now take just a few clicks — while keeping you in control and allowing visual inspection throughout the process.
To explore more features and examples, visit the full documentation here: https://documentation.samson-connect.net/tutorials/smiles-manager/using-the-rdkit-smiles-manager/
SAMSON and all SAMSON Extensions are free for non-commercial use. You can download SAMSON at https://www.samson-connect.net.





