Molecular modelers often face the challenge of going beyond static structures to understand how biomolecules move and function. In the context of viral mechanisms, visualizing transitions between different conformational states can reveal valuable insights about infection mechanisms and antibody binding sites. One pressing example is the SARS-CoV-2 spike protein, the molecular ‘key’ the virus uses to infiltrate human cells.
In this post, we’ll explore how SAMSON helps modelers visualize and better understand the dynamic opening motion of the SARS-CoV-2 spike protein—from closed to open conformation—based on data derived from PDB structures and computational interpolation tools. Whether you’re developing therapeutics or just exploring viral mechanisms, this animation of molecular motion can provide useful perspectives.
Why the Spike Motion Matters
The spike protein plays a critical role in viral entry by binding to the human ACE2 receptor. However, this binding is only possible when the spike transitions from a ‘closed’ to an ‘open’ conformation. Visualizing this process reveals how the receptor-binding domain becomes accessible. This insight is key to strategies such as vaccine design or antibody development because many neutralizing antibodies target these open conformations.
Closed to Open: What It Looks Like
Using SAMSON, researchers have created animated trajectories that show this viral spike transition. These animations are computed using interpolation between the PDB entries of the closed (PDB: 6VXX) and open (PDB: 6VYB) states.
Here are some views from the computed spike transition:
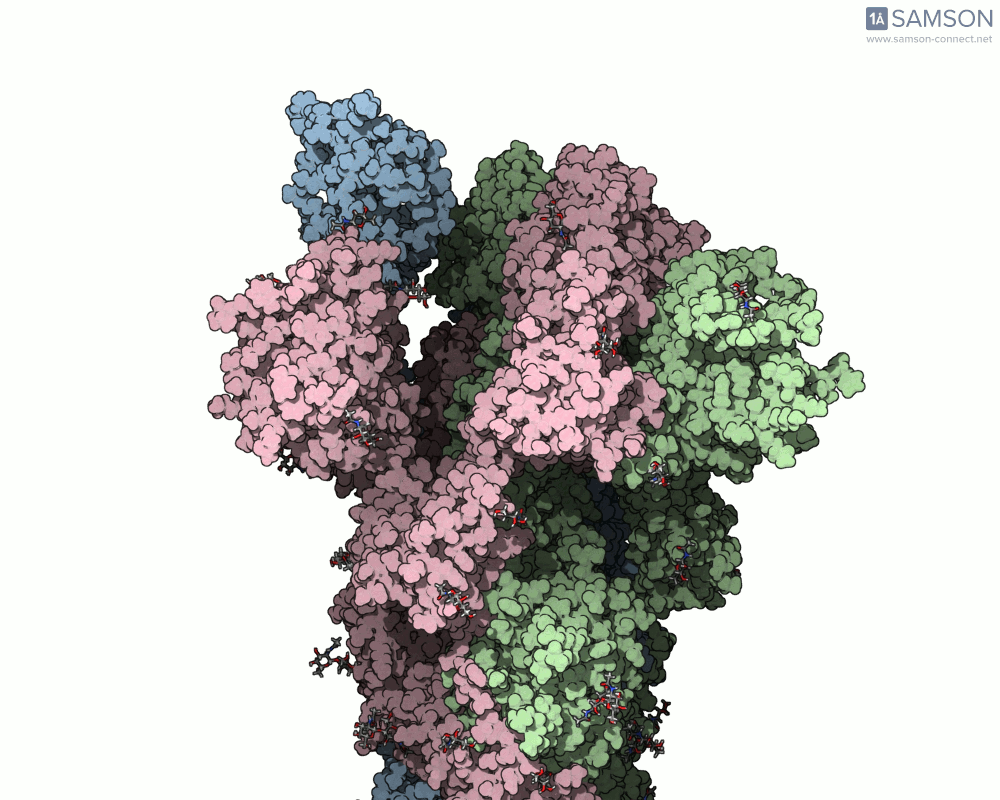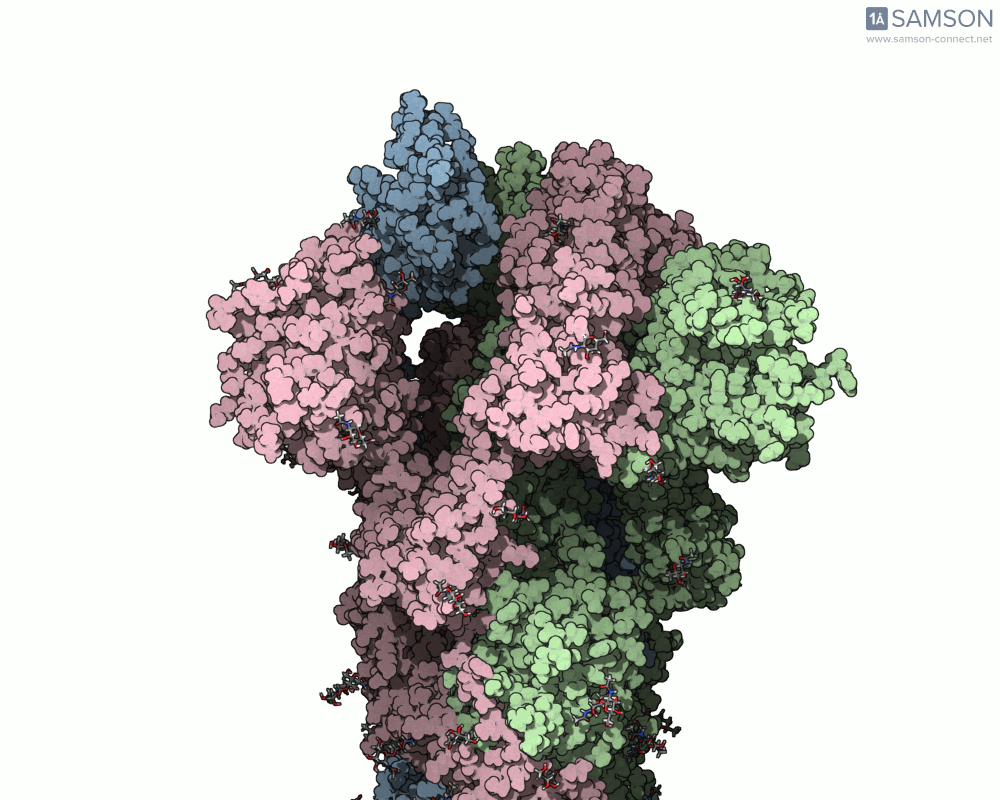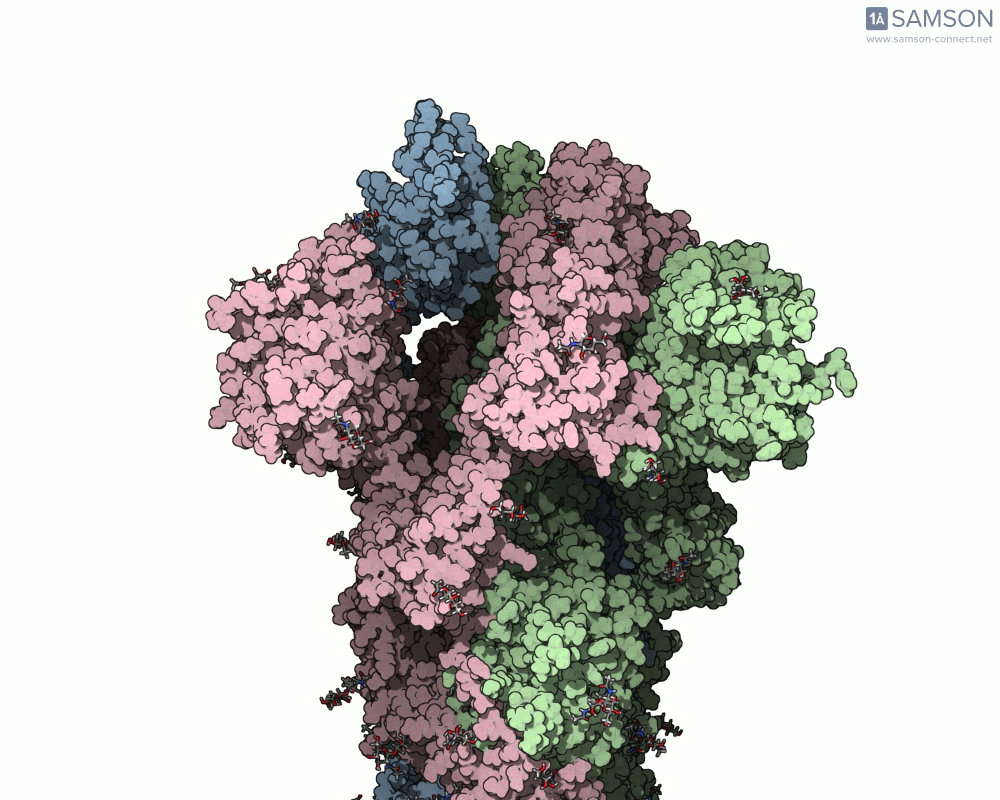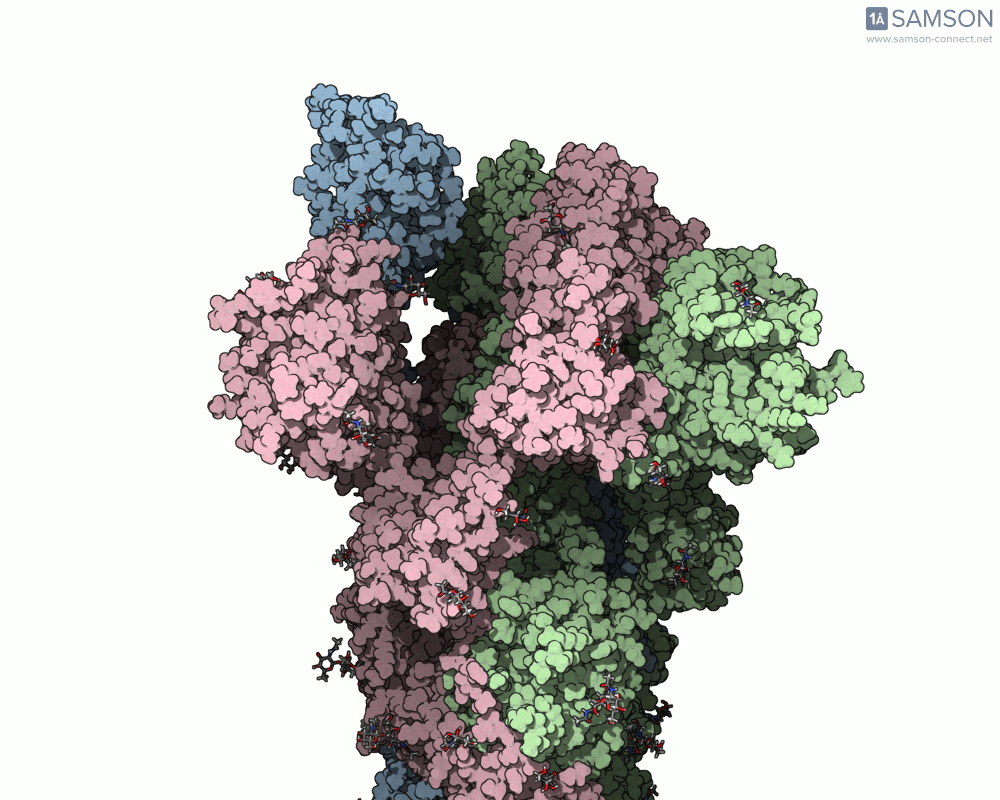
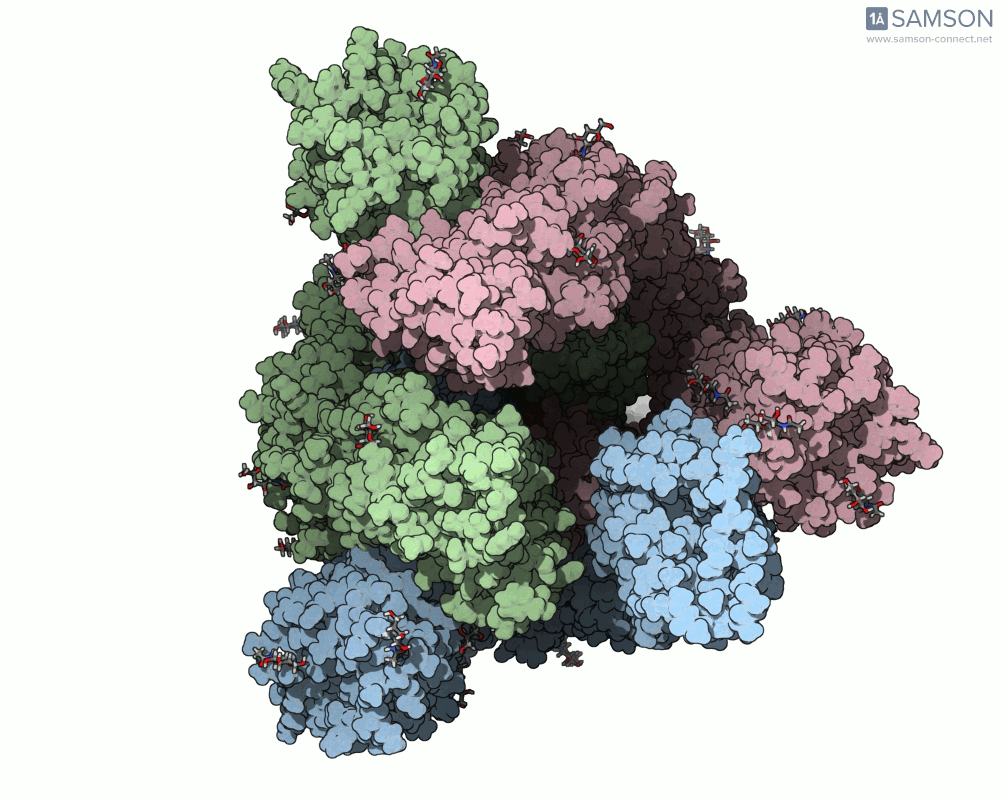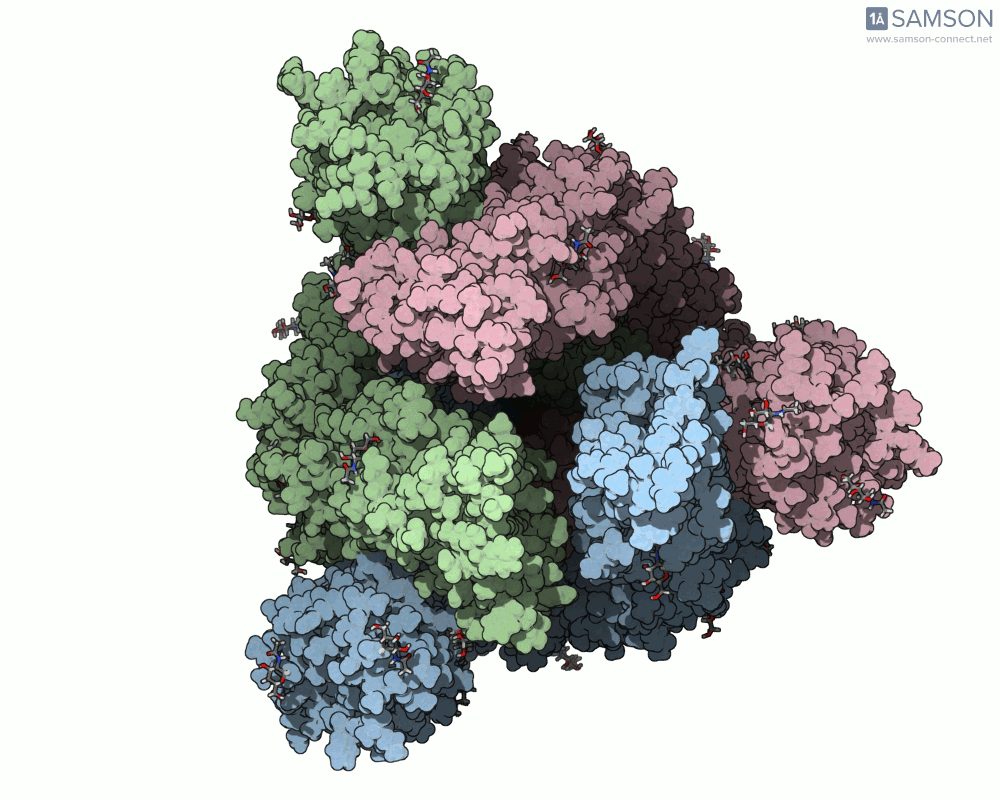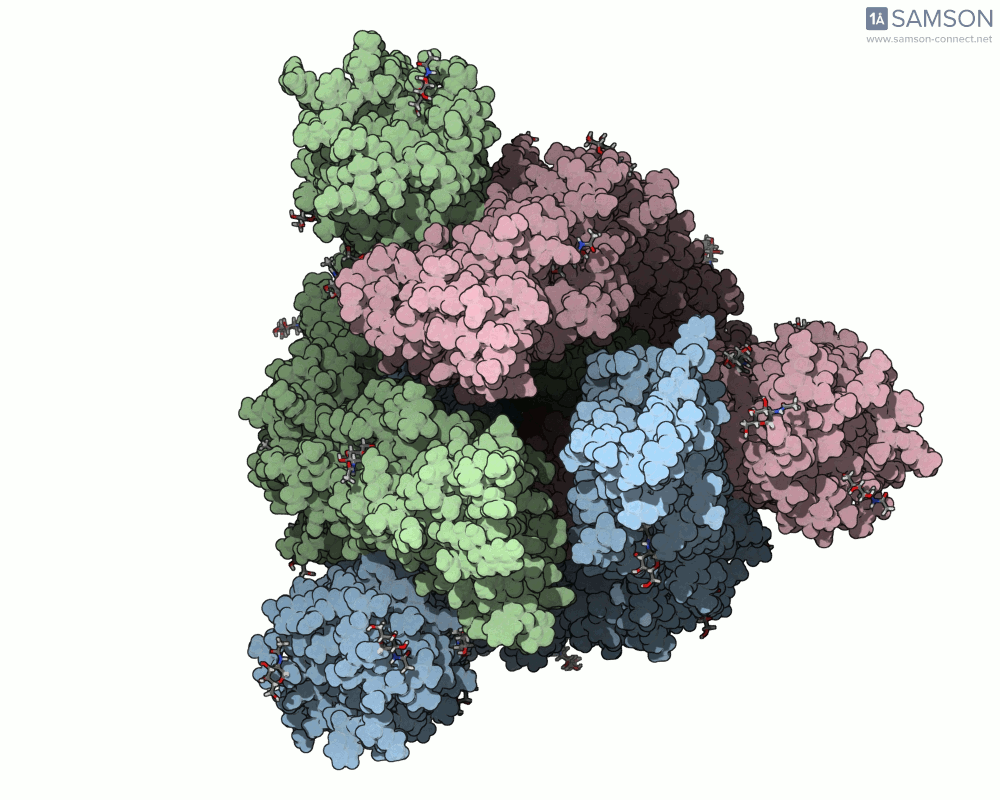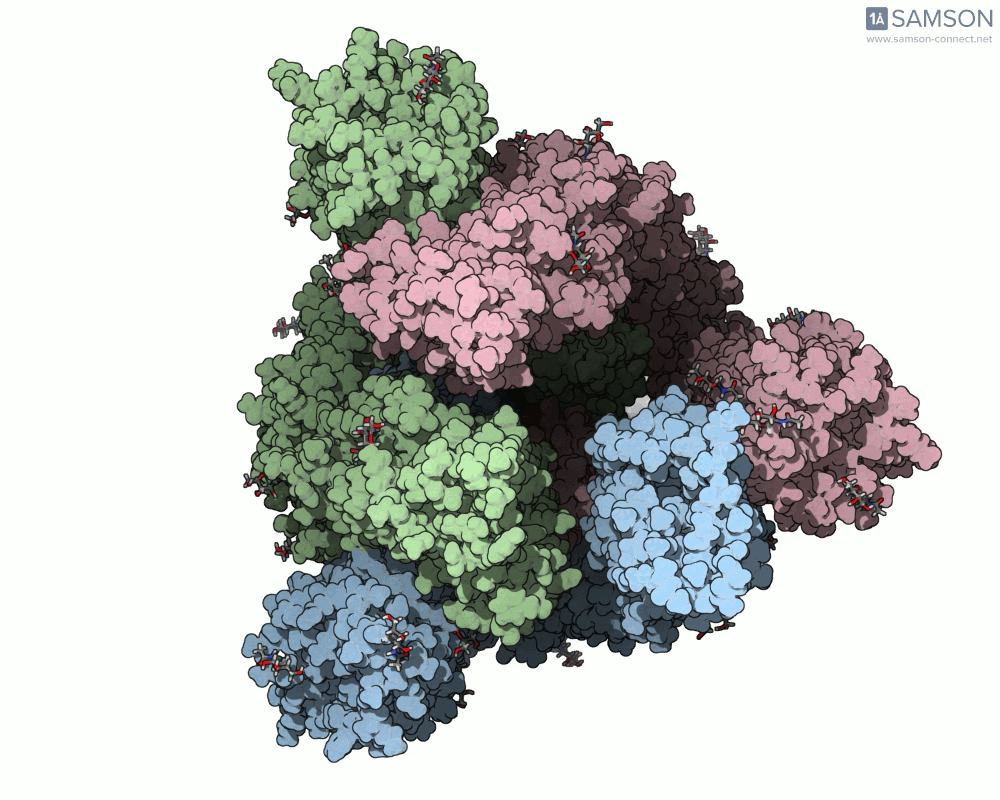
These animations are more than just visual aids—they help to hypothesize how the virus interacts with the host and suggest which angles or epitopes are exposed and potentially vulnerable to binding.
Behind the Scenes: How Was It Done?
The animation was generated in SAMSON through this workflow:
- Preparation of the open and closed spike structures (adding hydrogens, adjusting sugars).
- Generation of an interpolated path using the ARAP (As-Rigid-As-Possible) Interpolation Path module.
- Path refinement using the P-NEB (Parallel Nudged Elastic Band) module to obtain a more physically realistic transition.
The entire process was carried out on a standard laptop, showing that complex molecular motion can be modeled without requiring heavy computational resources.
Download, Inspect, Modify
If you’re curious to explore on your own, the spike motion trajectories can be downloaded in several formats:
The SAMSON file not only includes the spike structure but also the calculated path and pre-annotated states. You can double-click to toggle between the conformations or to control the animation.
More Than Just a Movie
If you’re developing models of viral neutralization or docking studies using the open conformation of the spike, seeing the motion gives you a more comprehensive context. It can also inform your hypotheses about transition states or design inhibitors that stabilize one conformation over another.
To learn more and explore the complete protocol, visit the original documentation page.
SAMSON and all SAMSON Extensions are free for non-commercial use. You can download the platform from https://www.samson-connect.net.





