When studying ligand exit pathways, molecular modelers often need a clean export of just the part of the system that matters: the ligand moving along its path. Exporting the full atom trajectories of an entire protein-ligand complex can quickly create large files that are cumbersome to process and visualize. If your goal is to extract snapshots of ligand movement over time—whether for enhanced sampling, animations, or free energy analysis—exporting just the ligand atoms offers a cleaner, more targeted solution.
Luckily, the Export Along Paths extension in the SAMSON molecular design platform gives you exactly that capability.
Why export a subset of atoms?
Consider this: you’ve discovered a viable exit path for a ligand using the Ligand Path Finder extension. You’ve optimized the path using a method like parallel nudged elastic band (PNEB), and now you want to run a free energy profile along this trajectory. You don’t need the entire protein structure at every frame, just the ligand coordinates as it progresses along the path.
This is where exporting a subset of atoms—rather than everything—saves time and resources.
Step-by-step: Exporting just the ligand
The Export Along Paths app allows precision control over what you export. Here’s how to define and export only your ligand:
- Open the app via Home > Apps > All > Export Along Paths.
- Expand the Advanced panel to work with groups of atoms.
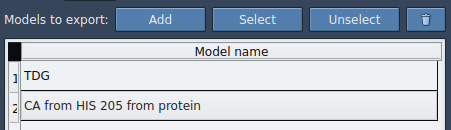
- In the Document view, select your ligand (e.g.,
TDG). - Click Add to define this molecule as a model you want to export.

This step creates a row in the export table representing your ligand, which you can rename (e.g., to ligand_only) and select or remove as needed.

You can continue to define additional groups of atoms—say, the binding pocket or protein backbone—for more complex exports.
Exporting the trajectory
- Select the relevant path(s) in the Document view.
- Choose whether all frames should go into one PDB file or be split across multiple files.
- Click Export atoms along paths to PDB files.
You’ll be prompted to choose the destination folder and a file prefix. The app handles the rest, saving a folder of ligand-only coordinates across the trajectory. This creates smaller and easier-to-analyze datasets—perfect for tools like GROMACS, NAMD, or VMD.
Applications
- Generates clean coordinate files for reaction coordinate profiling.
- Saves only the atoms you’re studying for enhanced sampling simulations.
- Creates simpler input files for free energy calculations or transition state analysis.
- Simplifies visualization of ligand movement.
Working with specific atom groups can significantly streamline your modeling workflow, especially when dealing with large biomolecular complexes.
To learn more, check out the full documentation here: Export atom trajectories along paths.
SAMSON and all SAMSON Extensions are free for non-commercial use. You can download SAMSON from https://www.samson-connect.net.





