When working with molecular libraries, identifying subsets of molecules that contain specific functional groups can be both repetitive and error-prone. Whether you’re hunting for fluorinated compounds, analogs with nitrogen-containing backbones, or custom motifs for SAR analysis, manual filtering isn’t scalable. This is where the SMILES Manager in SAMSON becomes valuable.
The SMILES Manager offers a feature-rich substructure search functionality powered by RDKit. This feature allows molecular modelers to quickly isolate and examine molecules that share specified chemical features based on their SMILES (Simplified Molecular Input Line Entry System) codes.
Why Substructure Search Matters 🔍
Imagine you’re working with a dataset of hundreds of SMILES and you’re interested only in those containing both a fluorine atom and a nitrogen atom in distinct parts of the molecule. Manually examining structures would be inefficient at best and error-prone at worst.
The substructure search feature provided by the SMILES Manager accelerates this task, allowing for more informed decisions during compound screening, functional group analysis, or preparing datasets for further modeling steps like docking or dynamics simulations.
How to Perform Substructure Search in SMILES Manager
1. Load your SMILES strings into the Manager (from .smi, .txt, or manually).
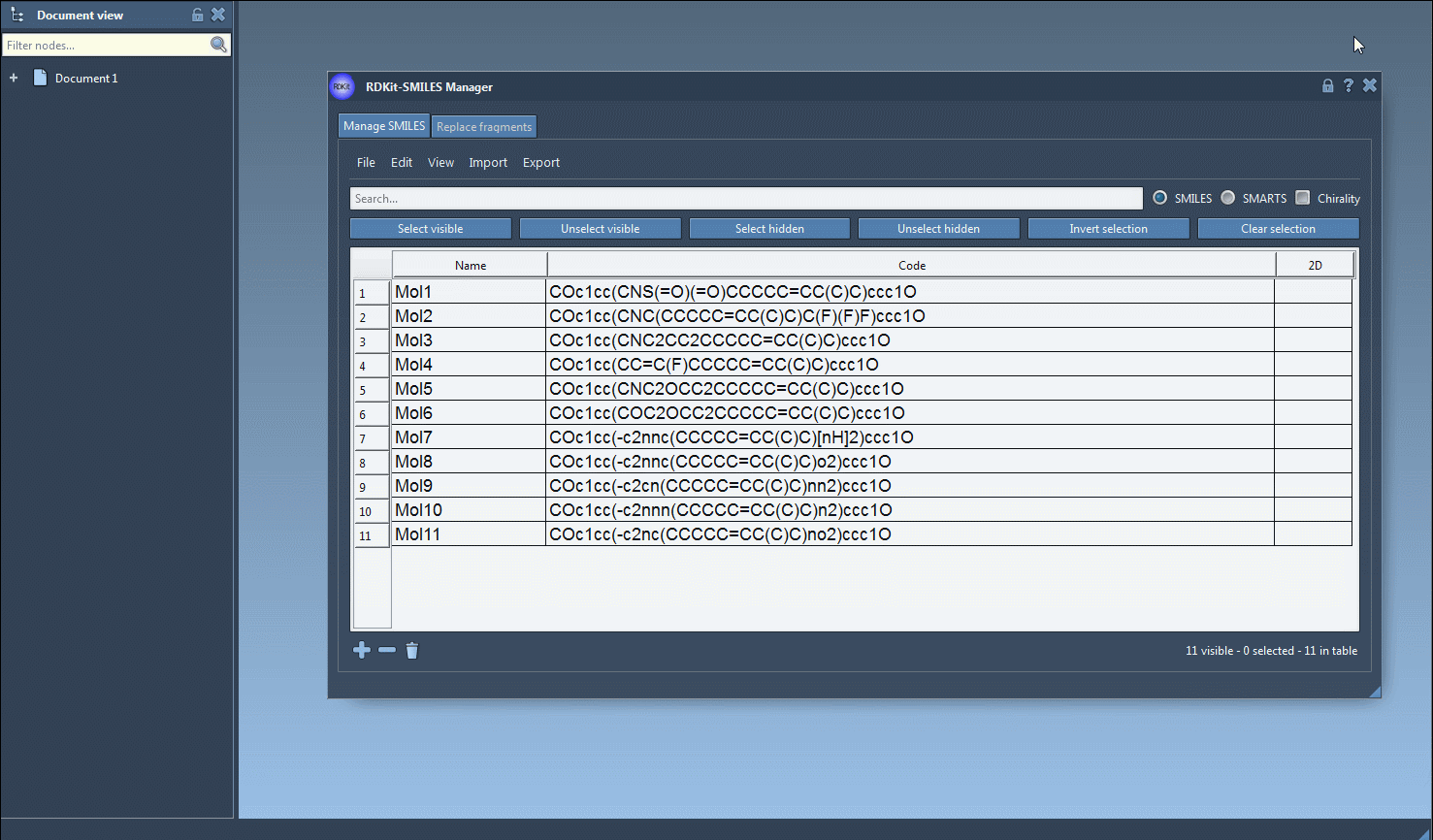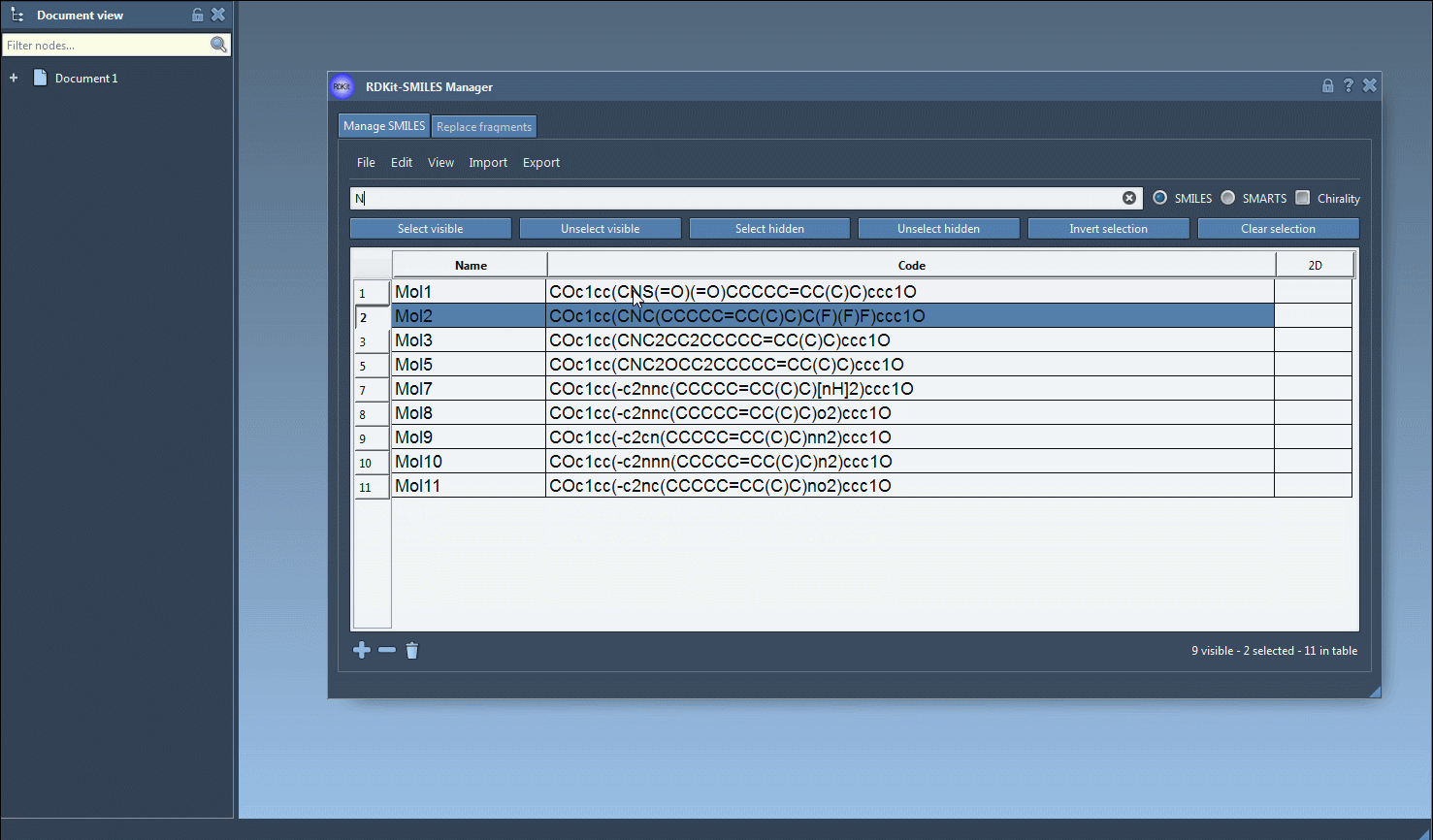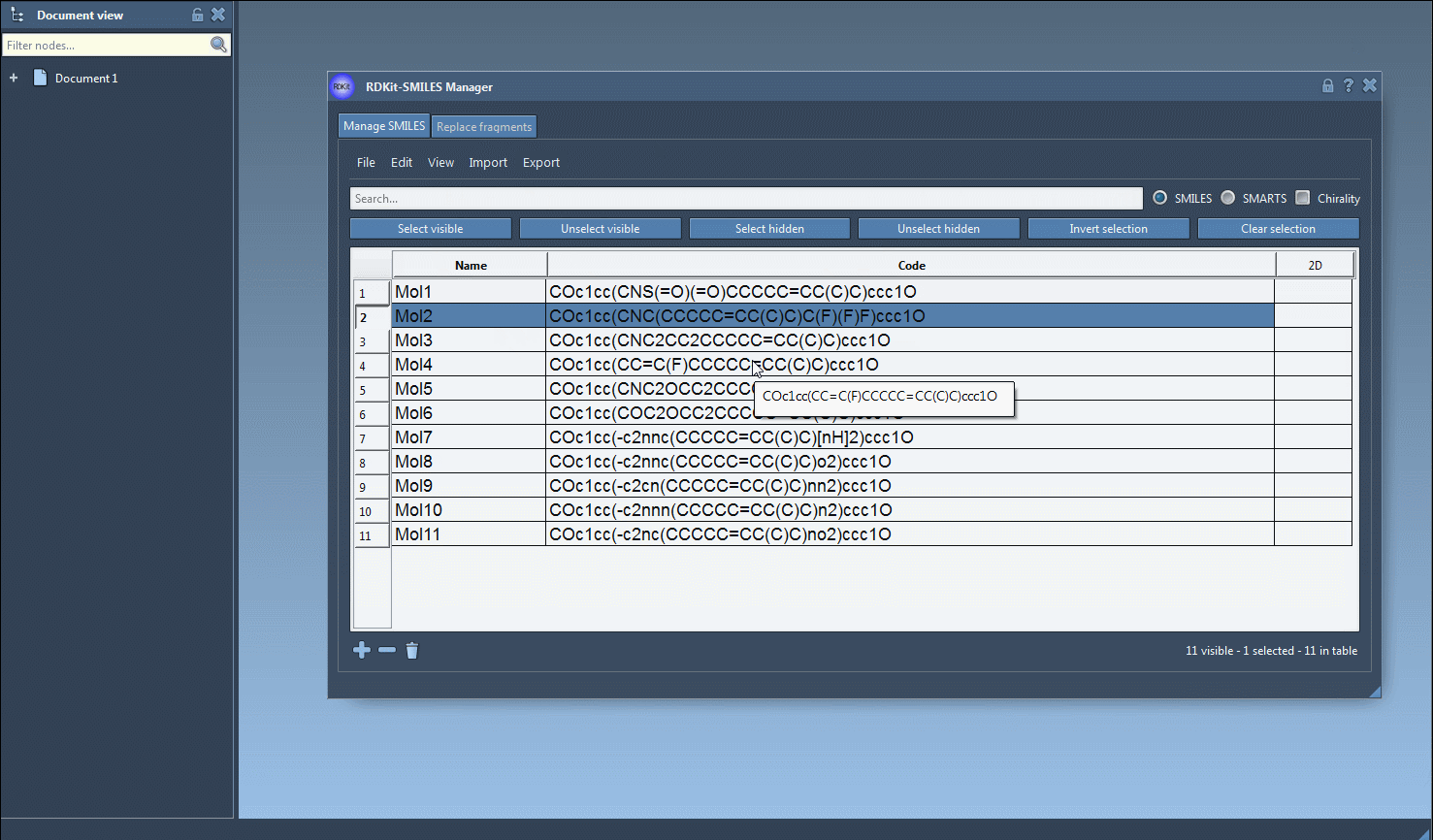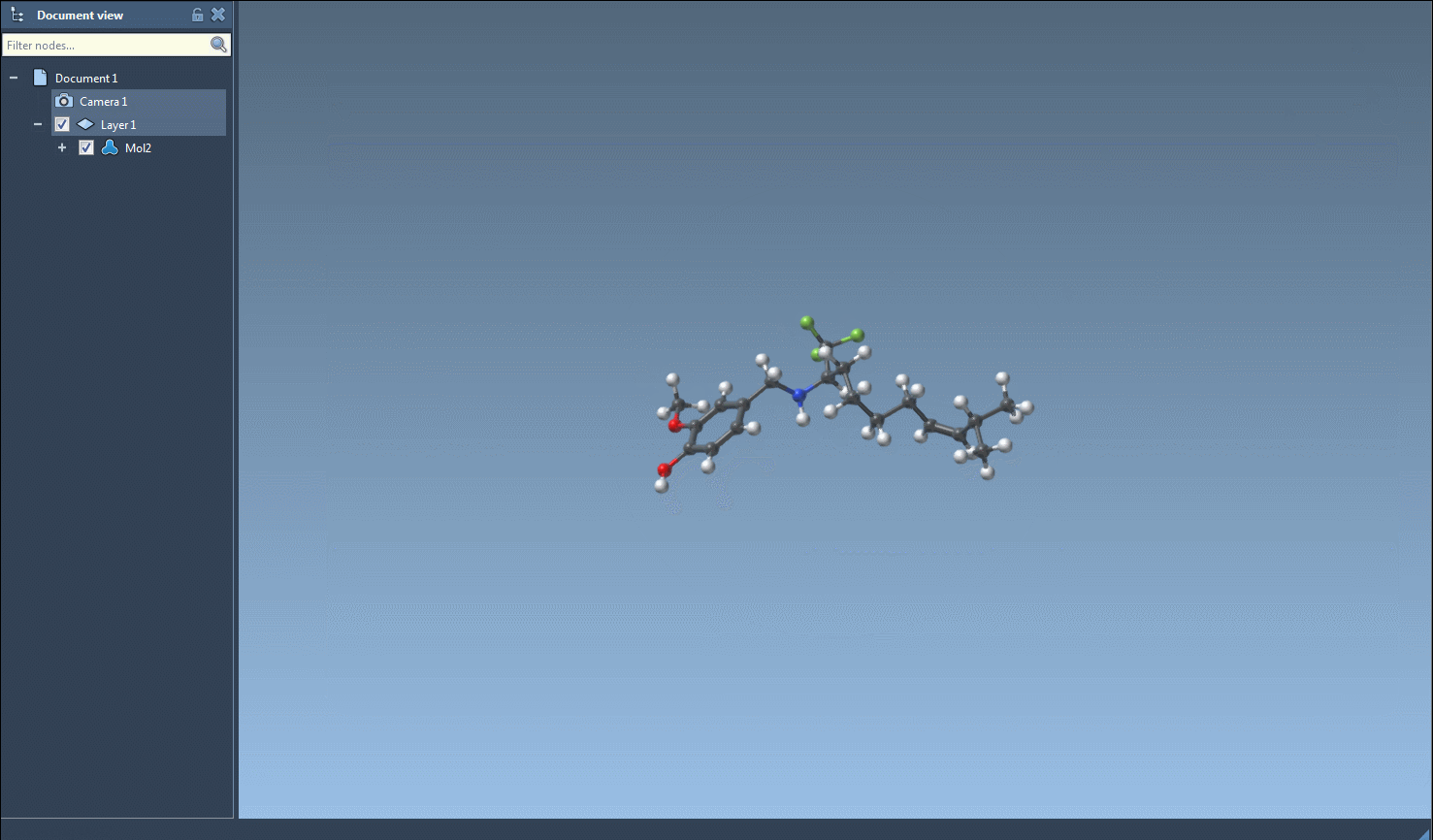
2. Use the Search bar to target a functional group or atom — for example, type F to search for molecules containing fluorine.
3. Then click Select visible to filter in only the visible (matching) molecules.
4. Now search for another group — such as N for nitrogen — and click Unselect hidden to further narrow the selection to molecules that contain both fluorine and nitrogen.
This stepwise logical operation lets you zero-in on molecules with precise structural characteristics — a key advantage during early-stage screening or lead optimization.
Visually, this is a highly intuitive process. The interface highlights matching entries and lets you perform all actions with point-and-click efficiency:
Tip: In RDKit, chirality elements are not considered in substructure searches by default. If you need stereochemical specificity, chirality options can be enabled.
Applications
- Target Identification: Find analogs with shared scaffolds or functional groups.
- Lead Optimization: Discover off-target features by isolating shared fragments.
- Data Cleansing: Remove undesired structural motifs prior to calculations.
By integrating substructure search directly into a visual molecular management tool, SAMSON makes it easy to connect chemical intuition with structured filtering logic — no scripting required.
Learn more about the SMILES Manager: documentation.samson-connect.net
SAMSON and all SAMSON Extensions are free for non-commercial use. You can download SAMSON at www.samson-connect.net.





