For many molecular modelers, the ability to rapidly identify molecules with specific features from a database is essential. Whether you’re looking for potential leads with fluorine groups for binding studies, or aiming to locate analogs containing nitrogen functionalities, the substructure search is often the lifeline to narrowing down a massive dataset.
But traditional substructure search tools often require switching between environments, exporting/importing files, or writing complex queries—interrupting the flow of design, analysis, and iteration. Enter the SMILES Manager in SAMSON, the integrative molecular design platform. One particularly helpful feature? A simple, visual interface for substructure searches powered by RDKit.
Start with a Visual Table of Molecules
The Manage SMILES tab presents your molecules in a sortable, editable table complete with names, SMILES codes, and 2D depictions. This alone already helps you keep track of what’s in your dataset. But here’s where things get interesting: with the search bar and selection tools, you can quickly filter, combine, and narrow candidates based on structural patterns you’ve defined.
Refining Your Query in Two Steps
Let’s say you’re within a dataset of drug-like molecules and want to find entries that contain F (fluorine) and N (nitrogen), but not necessarily adjacent to one another. Here’s how:
- In the search bar, type
F. Molecules that match will remain visible. Click on Select visible to select them. - Clear the search bar. The previously selected fluorinated molecules remain selected.
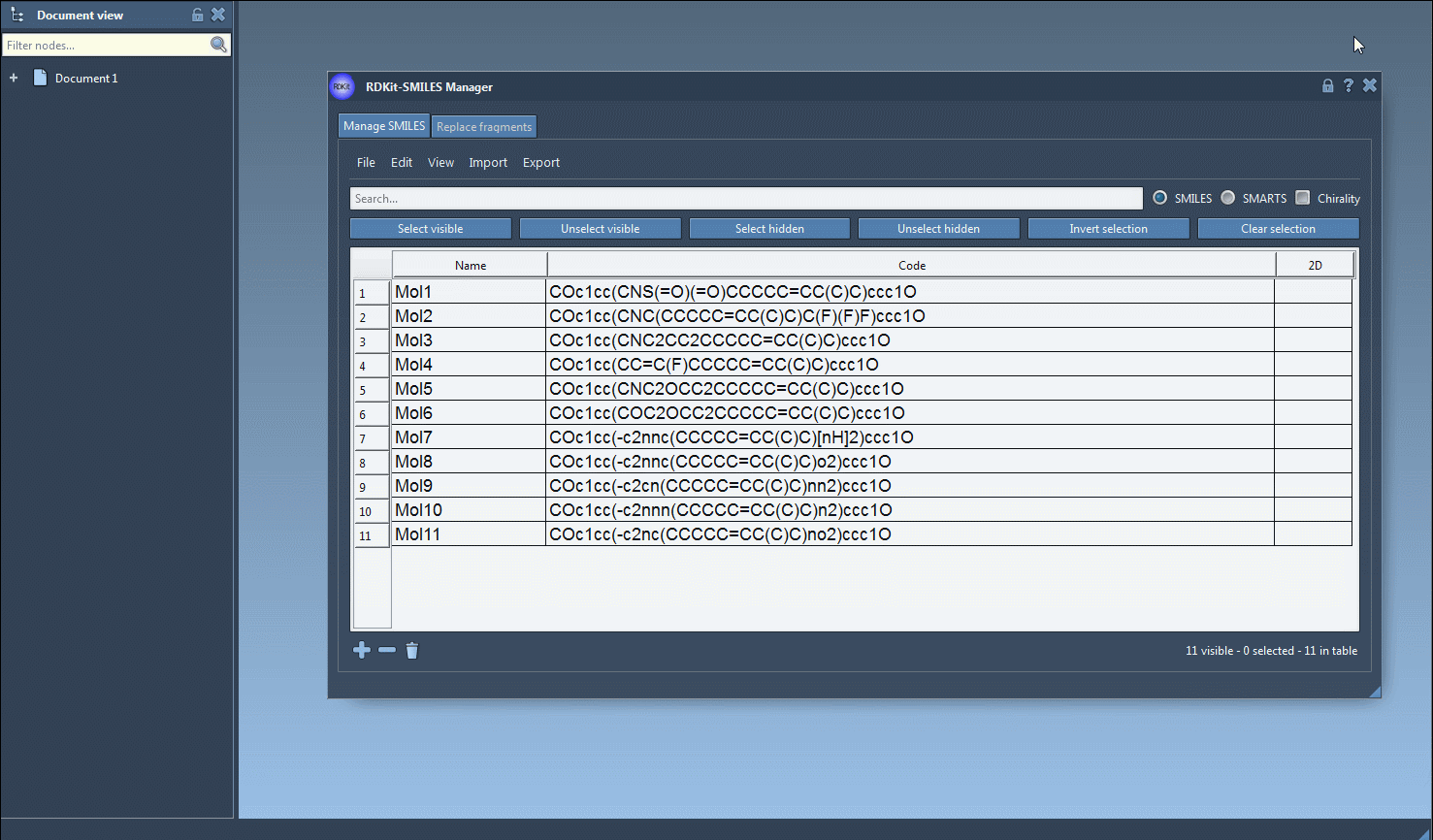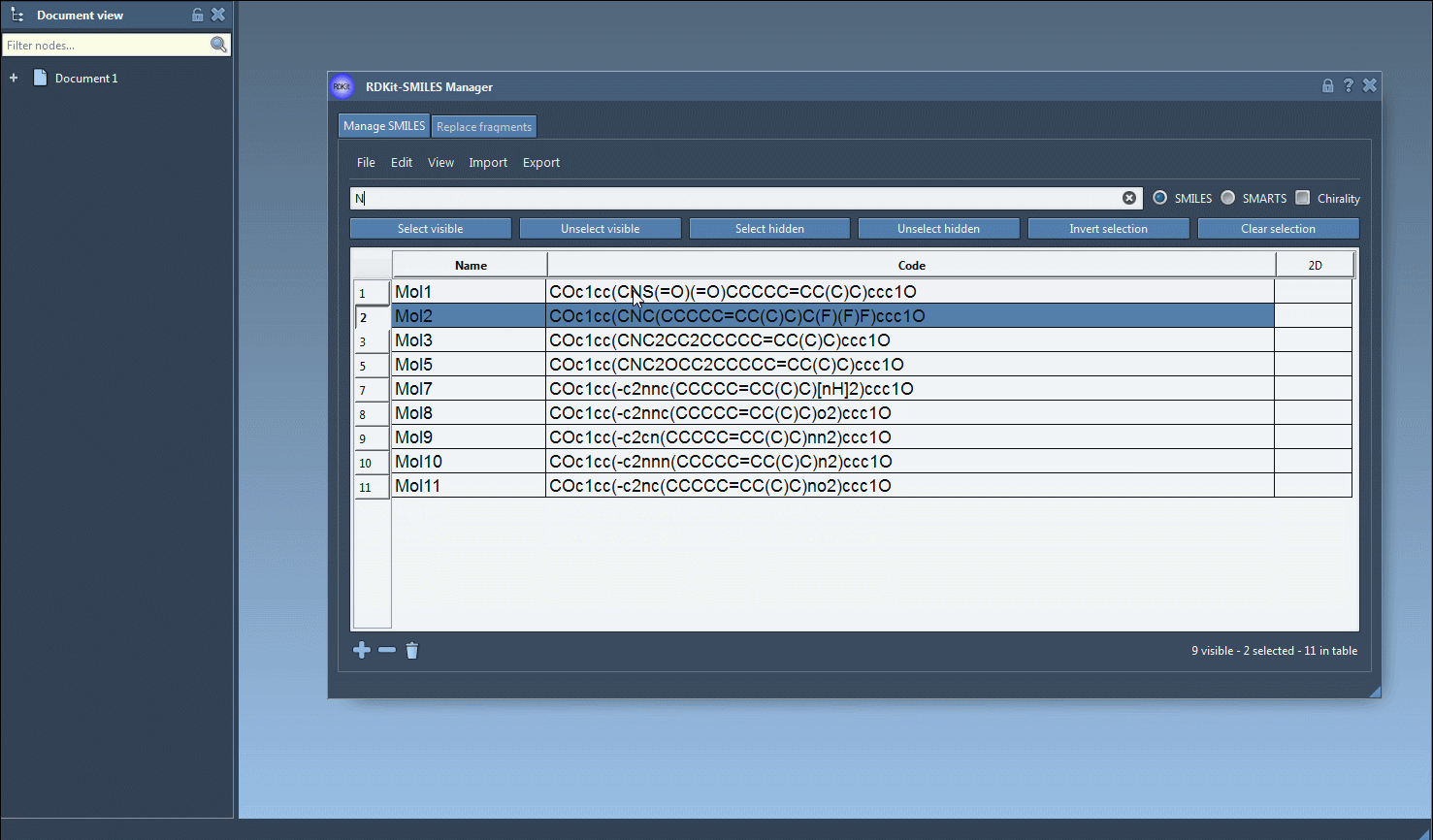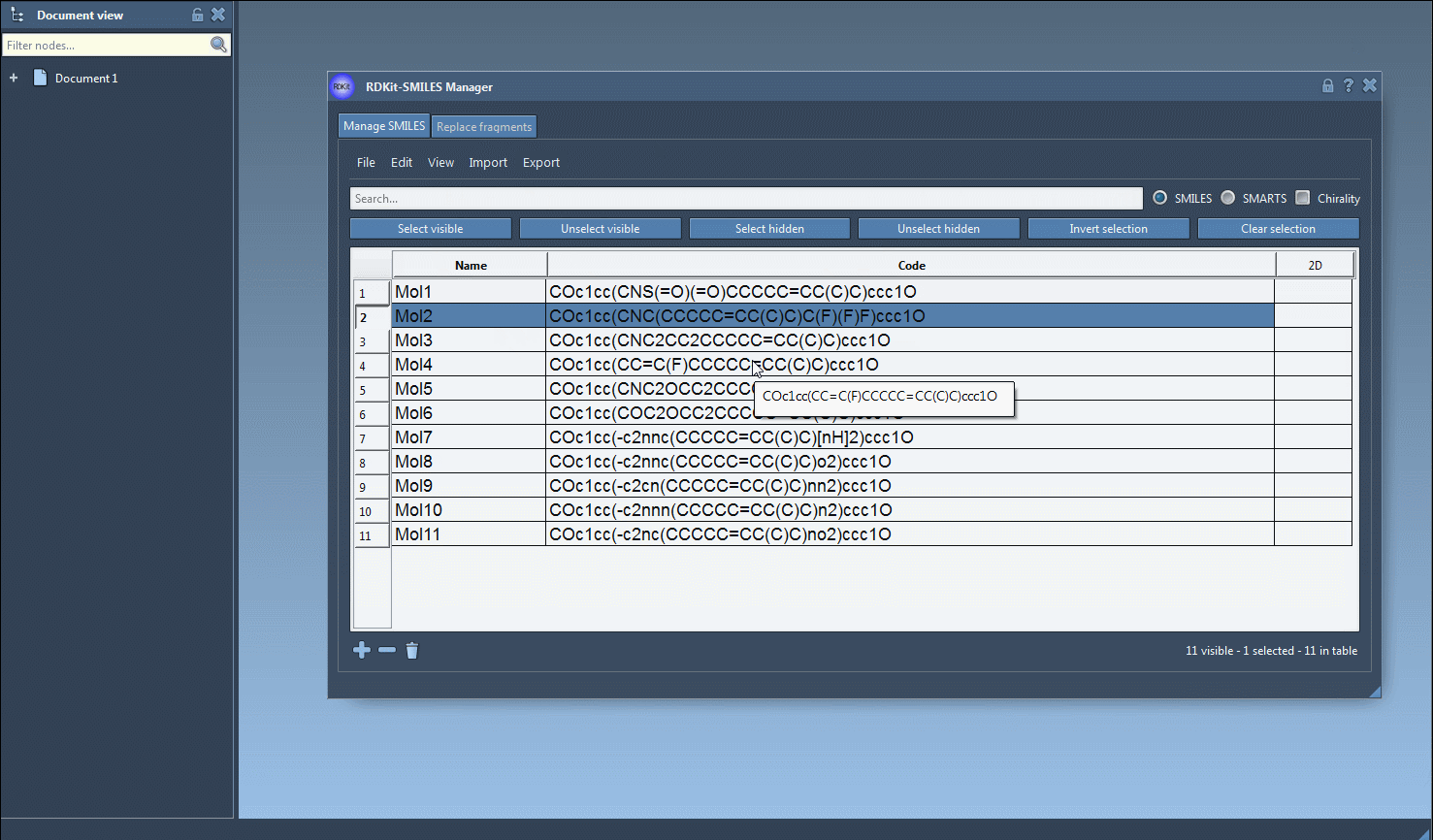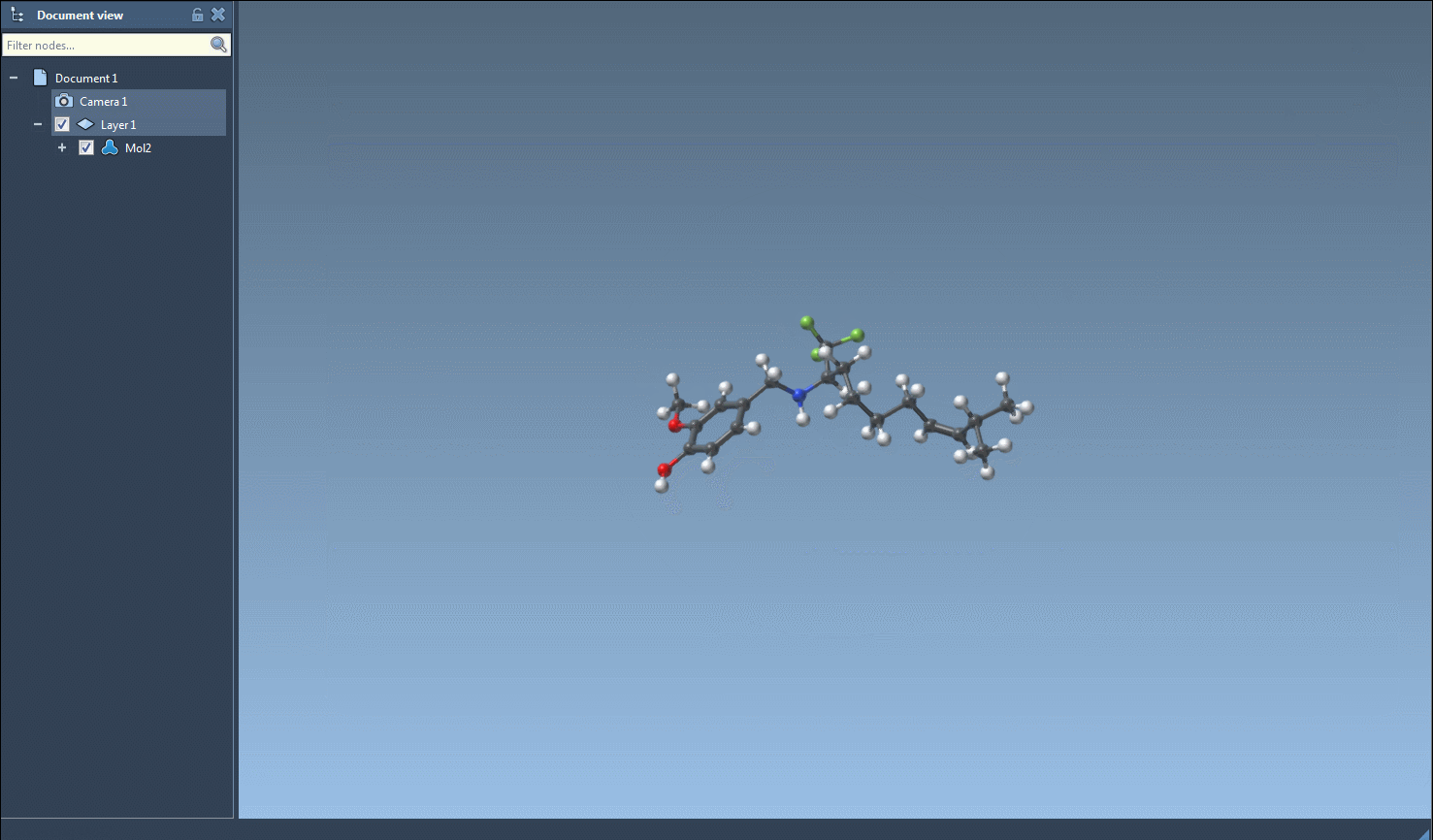
- Now search for
N. You’ll see how many of the already-selected molecules are also visible (i.e., contain nitrogen). - Click on Unselect hidden to remove any molecules without nitrogen from your selection.
After just these few clicks, you’re left with molecules containing both F and N. It’s a powerful yet user-friendly way to isolate candidates for further testing or modeling.
Useful Notes for Precise Filtering
- Searches are based on string matching with RDKit. You are not limited to elements—search for fragments, tautomers, or even stereoconfigurations.
- By default, stereochemistry is not considered. You can enable chirality if you need to differentiate between stereoisomers during the search.
- Combining with the table’s visual elements (e.g., 2D depiction previews) allows instant contextual feedback when decisions need to be made on-the-fly.
When Would You Use This?
If you’re performing medicinal chemistry optimization, preparing a virtual screening campaign, or simply organizing your molecular database, this substructure search saves time and removes friction from your workflow. You don’t need to export anything or write scripts—everything happens inside the same interface where you’re already working.
Learn more about SMILES Manager in the full documentation.
💡 SAMSON and all SAMSON Extensions are free for non-commercial use. Get your copy at https://www.samson-connect.net.





