Creating complex polymers often involves repeating monomer patterns, varied bond types, and the need for flexible editing. For many researchers and students in the molecular modeling field, building such custom polymer sequences can feel intimidating — especially when it involves command-line tools or scripting. But what if you could do it interactively, without writing any code, and with full visual control?
This post focuses on a specific feature available in SAMSON’s Polymer Builder: the ability to register monomer sequences and use them to build complex polymers. It addresses a common challenge in polymer modeling — defining structured, reusable subunits in your macromolecule — and presents a no-code solution that could save you time and reduce errors.
What’s a monomer sequence?
A sequence is a set of registered monomers, identified by automatically assigned letters like A or B, arranged in a specific order (e.g., ABBA). These sequences define how monomers connect, and you can assign them bond types (single, double, triple) using symbols like = and # (e.g., A=B or A#B). Named sequences like S1, S2, etc., can then be combined to build larger polymers.
This approach helps you:
- Capture repeating units clearly, which is useful for block copolymers or alternating sequences.
- Avoid re-entering the same monomer strings over and over again.
- Assign clear identifiers for sequences that can be reused, modified, or combined.
How to get started
Once you have registered your monomers (each represented by a letter), switch to the sequence registration section in the Polymer Builder interface. To define a new sequence:
- Click Add new sequence.
- Enter the sequence using your monomer IDs (e.g.,
ABBA). - Optionally assign connectors like double or triple bonds (e.g.,
A=B,B#A). - Give your sequence a name if you’d like it isolated in your final polymer structure.
The system checks the validity of the sequence and immediately flags errors in the Status column — helpful when experimenting with complex arrangements.
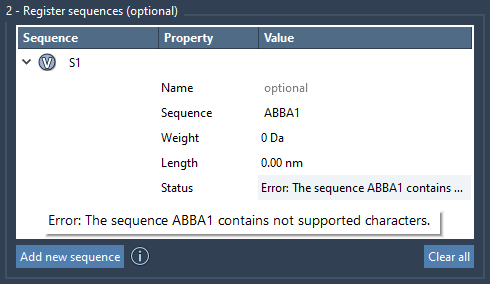
Visual and editable
After registration, sequences appear in a table along with their approximate molecular weight and length. This feedback helps estimate the polymer’s size without full construction. Highlighting which monomers are involved in each sequence is just a click away (by pressing the V button).
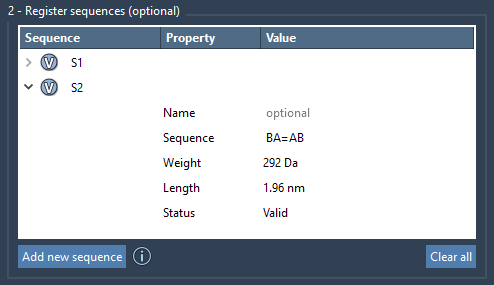
You can easily update or rename a sequence in the table, and if needed, remove it by right-clicking it and selecting Delete sequence. Similarly, you can clear all sequences in one go with Clear all.
Why it’s worth using
This structured approach is helpful for users interested in constructing polymers with specific architecture — whether it’s for property prediction, coarse-grained modeling, or experimental setups. It’s especially valuable when you are testing various hypotheses and want to quickly adjust your design.
Rather than building everything manually, storing sequence information gives you repeatability, precision, and rapid iteration.
To learn more about defining and using sequences in the Polymer Builder, visit the original documentation page.
SAMSON and all SAMSON Extensions are free for non-commercial use. You can get SAMSON from https://www.samson-connect.net.





