Sometimes, even with a promising homology model, everything just looks right—until you dig into the backbone geometry. Strained or disallowed dihedral angles can hide within your structure, only to show up later in your simulations as instability or poor predictive results. This is a common pain point in protein modeling workflows, especially when preparing structures for dynamics or design.
One surprisingly intuitive way to improve your structural models is to use a combination of non-linear normal modes and an Interactive Ramachandran Plot. Within the SAMSON platform, this combo not only helps you visualize problematic residues, but also allows you to refine them interactively—all in 3D space.
Why Normal Modes?
Normal Mode Analysis (NMA) lets you describe large-scale movements in a biomolecule. These are the ‘soft’ motions proteins use for things like allostery or domain movements. When applied smartly, they provide conformational variety while staying within energetically reasonable paths.
By combining NMA with Ramachandran visualization, you can fix local backbone issues using a global motion context—meaning changes look more natural and biologically plausible.
A Step-By-Step Mini Tutorial
- Load a protein structure, like PDB ID
1YRFin SAMSON via Home > Fetch. - Activate the Interactive Ramachandran Plot by installing the corresponding Extension and opening the app in Home > Apps > Biology.
- Check your model: Click Update in the Ramachandran Plot app, and examine whether residues are in allowed regions (yellow = good, white = problematic).
- Activate Normal Mode Analysis:
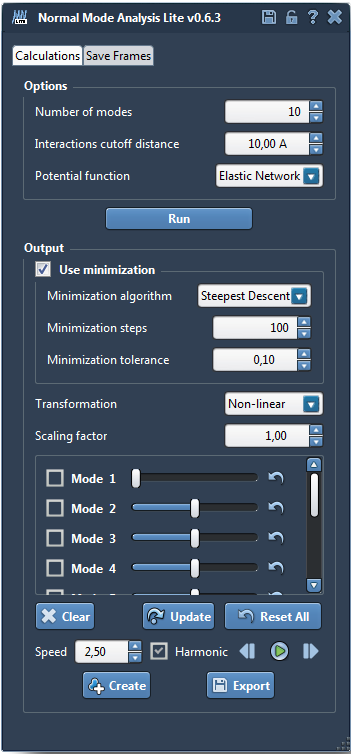
- Compute the first 10 non-linear normal modes of your structure (see SAMSON’s normal modes tutorial).
- Set the scaling factor to 1 and apply the first normal mode’s displacement by shifting the slider all the way to the left.
- Tweak with the Ramachandran Plot:
- Zoom into the plot and click on residues in the white (unfavorable) zones.
- Drag them into acceptable (yellow) areas and notice that the changes instantly update the 3D conformation.
Why This Workflow Is Worth Trying
This approach solves the common problem of local strain in protein backbones. Instead of fixing outliers residue-by-residue in an isolated way, using normal modes keeps the overall architecture intact while unlocking flexible refinements. It’s particularly useful when finalizing homology models, preparing for molecular dynamics simulations, or assessing conformational flexibility near active sites.
Combining SAMSON’s normal mode tools and the Interactive Ramachandran Plot creates a streamlined editing experience that doesn’t require scripting or starting over. It’s interactive, visual, and well-suited for researchers who want to validate and optimize without too much overhead.
This quick workflow is easy to include as a routine part of model cleanup—especially when transitioning from structure prediction to simulation or design.
To learn more, check the full tutorial: Interactive Ramachandran Plot Documentation.
SAMSON and all SAMSON Extensions are free for non-commercial use. Get SAMSON at https://www.samson-connect.net.





