When studying molecular systems—especially large biomolecules—understanding how ligands move, how proteins shift domains, or how groups of atoms migrate through a structure is often central to your investigation. Yet, translating this dynamic behavior from raw trajectories into visually meaningful representations can be a hurdle.
A common challenge: you’re running simulations and you know certain atoms are moving along interesting pathways. But visualizing this in an intuitive, shareable, and analyzable way? That’s harder than it should be. You want to see how a protein’s domain flexes, how a ligand exits a binding pocket, or just where a cluster of atoms goes. Not every motion needs atomic-level detail—sometimes the movement of the center of mass (COM) is all you need.
This post introduces a simple workflow using the Pathlines visual model in SAMSON, tailored for visualizing trajectories of a system’s center of mass. It’s a direct way to track molecular motion across various pathways—whether you’re studying ligand unbinding, diffusion, or domain shifts.
Why Use a Center of Mass Pathline?
Sometimes, following every atom is just too much detail. Tracking the COM is not only computationally lighter but yields clear trajectories that are immediately intuitive. The Pathlines visual model allows you to:
- Track the motion of selected atoms or residues along defined paths.
- Visualize unbinding and rebinding events of ligands.
- Get an overview of domain-scale movements in large proteins.
How It Works in SAMSON
Here’s how you can go from your molecular data to a COM trajectory visualization in just a few clicks:
1. Load Your System
Start by opening a model in SAMSON that includes molecular pathways. A good example is the sample system featuring Lactose permease and its ligand TDG, available via the download link:

2. Select Your Atoms and Paths
In the Document view, select a group of atoms (e.g., your ligand) and one or more paths. If you don’t select anything, the whole document will be used by default.

3. Create the Visual Model
To generate the pathline:
- Go to Visualization > Visual model > More…
- Select Pathline of the center of mass
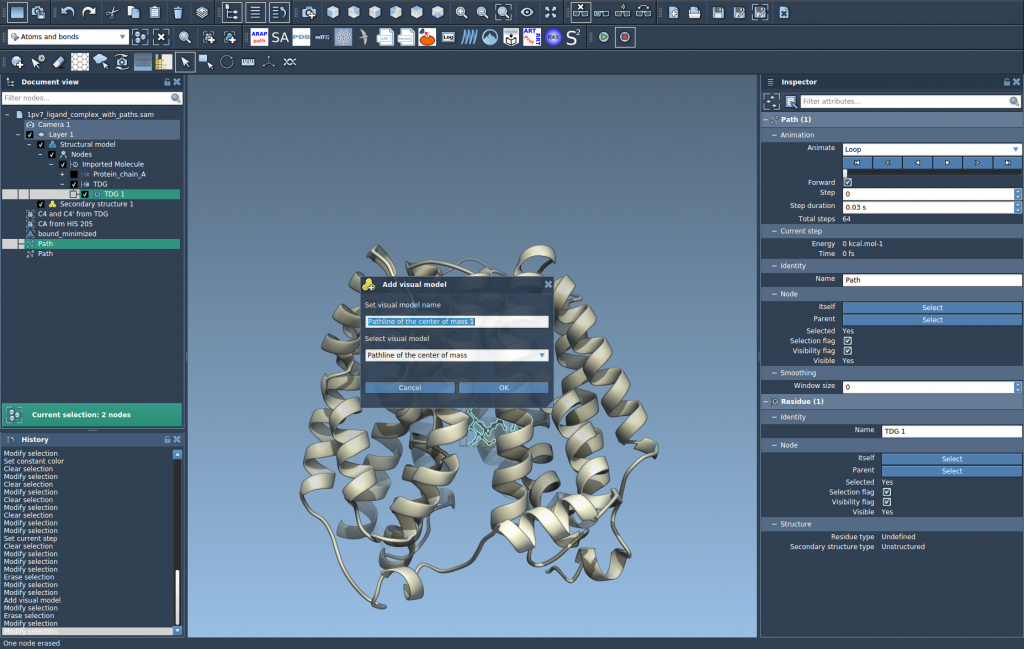
This creates a trajectory that shows how the COM of your selected atoms moves along the selected paths.
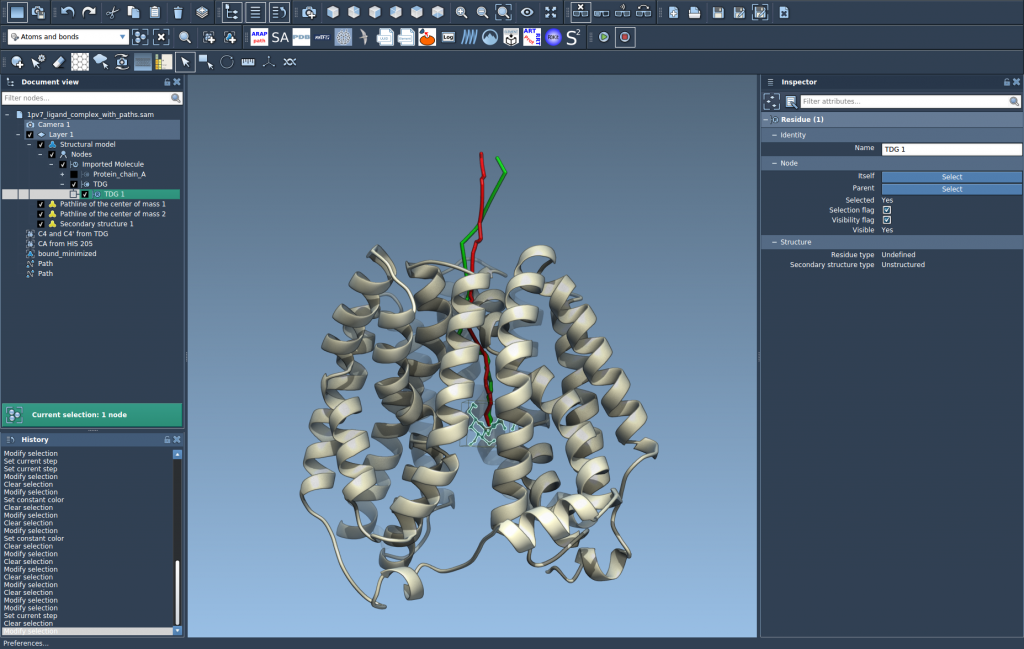
4. Explore and Tweak
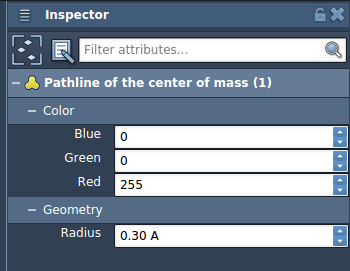
You can fine-tune visual aspects like color and thickness through the Inspector panel. Pathlines automatically update when paths or atoms are modified, and you can interactively control path playback or inspect individual points.
Use Cases
Whether you’re analyzing conformational changes or watching small molecules exit a receptor, knowing where the motion is going can be more important than how each atom rattles. Here are a few use cases:
- Displaying ligand unbinding or rebinding routes.
- Revealing collective domain movements in proteins.
- Monitoring COM shifts as a proxy for reaction coordinate progress.
This method is especially effective when paired with tools like Ligand Path Finder or P-NEB, which define paths based on energetic considerations. Visualizing the COM along such computed paths highlights the principles guiding molecular behavior.
To dive deeper into creating once you’ve visualized, check out the original tutorial page: Pathlines Tutorial.
SAMSON and all SAMSON Extensions are free for non-commercial use. You can download SAMSON at https://www.samson-connect.net.





