Understanding how proteins arrange themselves into larger structures is a recurring challenge in molecular modeling. Whether you’re studying quaternary structures or designing nanostructures, one issue always arises: how to easily visualize the full biological assembly from the asymmetric unit provided in a PDB file.
This is where the Symmetry Mate Editor in SAMSON can help. This tool lets you recreate symmetry mates directly from CRYST1 and BIOMT records in PDB files, giving you quick access to complete, realistic assemblies with just a few clicks. In short, it turns fragmented information into full 3D insight.
What Are Symmetry Mates?
When you download a PDB structure, you might not get the full biological assembly—the complete, functional form of the protein. Instead, you get just a part: one asymmetric unit. The PDB file often contains hidden transformation data that can reconstruct the entire biological or crystallographic assembly. The Symmetry Mate Editor uses these hidden records to automatically generate those meaningful assemblies.
CRYST1 vs BIOMT: The Two Symmetry Sources
The PDB format includes two main types of transformation records:
- CRYST1: Describes symmetry from the crystal lattice.
- BIOMT: Encodes biologically relevant transformations, such as quaternary assemblies.
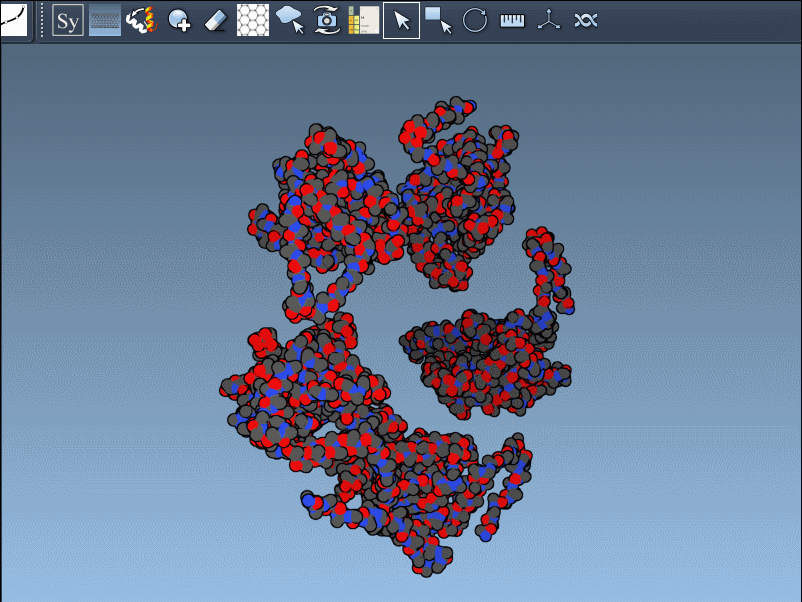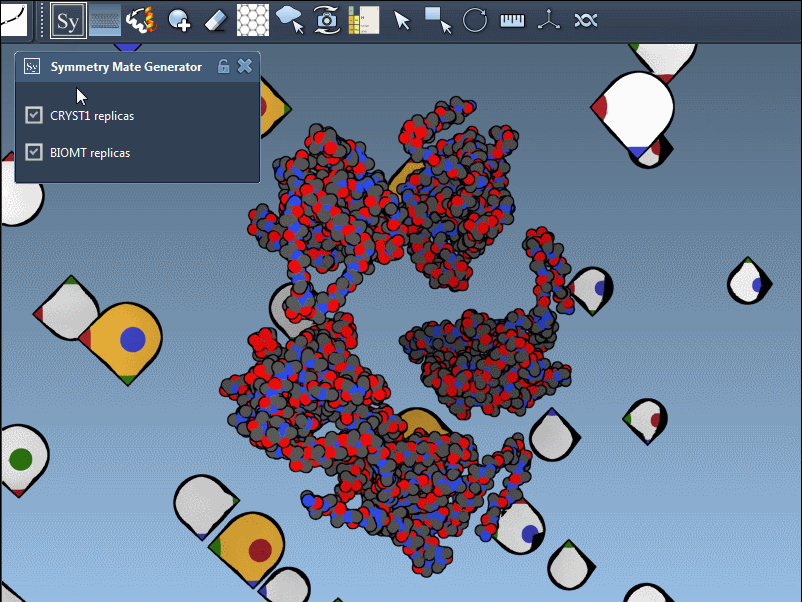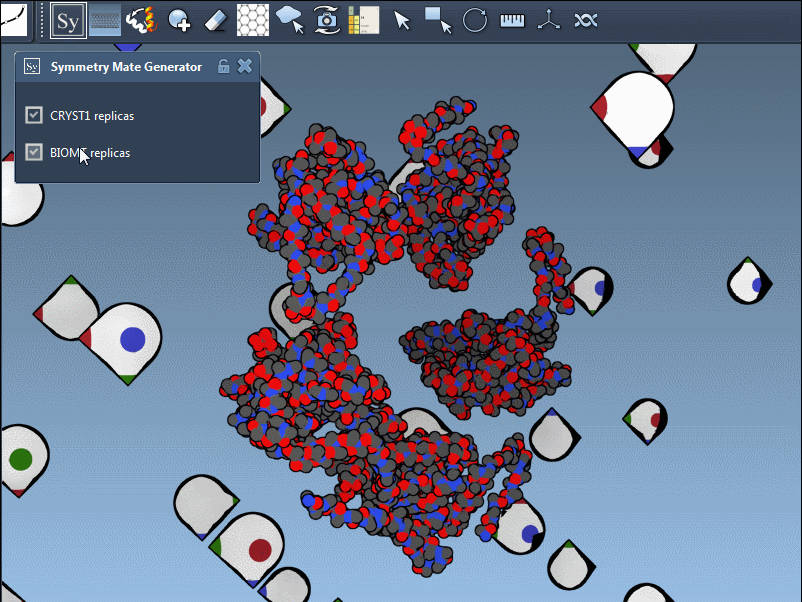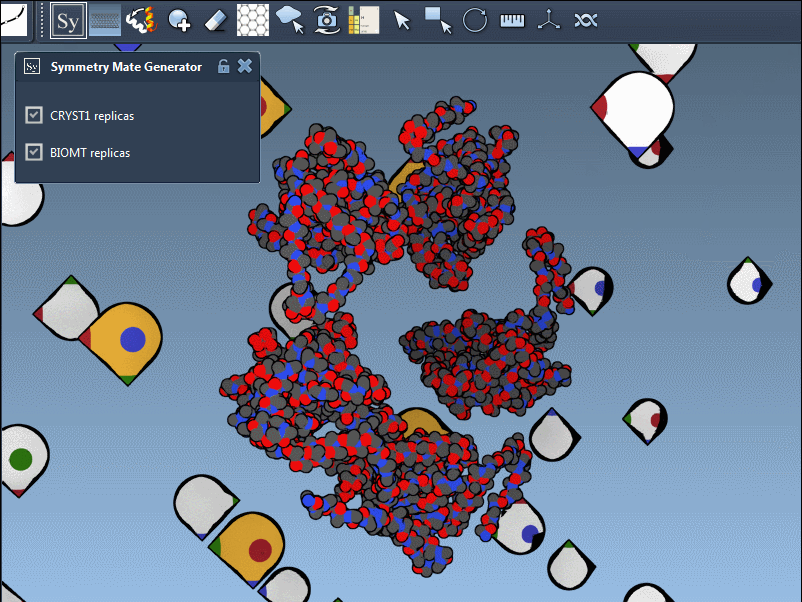
The Symmetry Mate Editor supports both and represents them visually in the viewport:
- White control nodes for CRYST1 records
- Yellow control nodes for BIOMT records
This lets you instantly know the origin of each suggested transformation. You can switch between the two directly in the editor interface—no scripting required.
Preview First, Generate Later (Or All at Once)
You don’t need to apply all transformations blindly just to see what they do. Hovering over a control node previews the potential symmetry mate in real time. Ready to keep it? A simple left click generates it permanently. Need to create all possible mates in one go? Hold Ctrl (or Cmd on macOS) before previewing and clicking.
This combination—preview before generation and control over type of transformation—is especially useful when working with crowded lattice structures or complex biological assemblies.
Real Applications in Molecular Modeling
Reconstructing full assemblies isn’t just for visualization—it’s often the first step in serious modeling work. Symmetry mates are crucial when:
- Preparing full oligomers for simulations
- Designing symmetric protein cages or scaffolds
- Testing repetitive binding site behavior across interfaces
- Simulating interactions at protein-protein or protein-ligand interfaces
By immediately recreating the real architecture of the molecule you’re studying, you reduce guesswork and eliminate the risk of modeling an incomplete structure.
Learn more from the documentation page.
SAMSON and all SAMSON Extensions are free for non-commercial use. You can get SAMSON at https://www.samson-connect.net.





