What to Do When You Have Conformations, But Not a Path?
Quickly Find the Best Binding Modes with the AutoDock Vina Extended Viewer
Zoom Without Wobble: How to Create Clean Zoom Ins in Molecular Animations
Choosing the Right Periodic Box Strategy for Batch Simulations in SAMSON
A Quick Way to Create Custom Index Groups for GROMACS Pulling Simulations
A Quick Way to Import an Entire NMR2 Tutorial Setup into SAMSON
Keep Your Molecular Views Consistent While Animating
Use the Disassemble Animation to Showcase Molecular Complexity
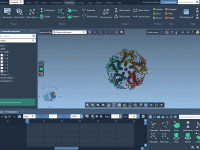
Creating compelling molecular animations can be surprisingly tedious, especially when presenting multi-component systems. A common challenge in structural bioinformatics and molecular modeling is illustrating how different components relate spatially in a complex supramolecular assembly. Whether it’s for teaching, a publication,…