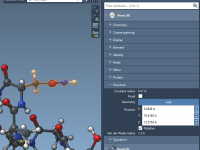
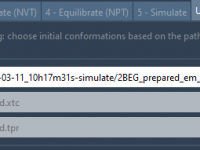
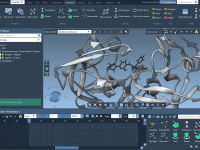
Setting Up Umbrella Sampling Projects From GROMACS Trajectories
Making Atoms Appear Progressively in Molecular Animations
When Structure Files Become Headaches: How SAMSON Helps You Parse What Matters
A Clearer View: Using Visual Models in SAMSON to Understand Nanosystems
Making Molecular Models Breathe with the Pulse Animation
Quick On, Quick Off: Using the Flash Animation for Clean Molecular Visibility Transitions
Creating engaging molecular animations is often essential when preparing scientific presentations, tutorials, or visualizations for molecular modeling projects. One recurring challenge is how to briefly yet clearly showcase the appearance and disappearance of molecular components—without overwhelming the viewer with effects…
Quickly Finding Conformations by Atom Count in SAMSON
Clarifying Molecular Differences with Diverging HCL Color Palettes in SAMSON
Visual clarity is essential when working with complex molecular models, especially when highlighting differences such as binding affinities, charge distributions, or simulation deltas. But using the wrong color scheme can cause confusion—or worse, misinterpretation of critical regions. If you’ve ever…