Making Molecular Models Fade into View with the Appear Animation
Previewing Protein Symmetry Mates Without Overloading Your Workspace
Tired of cluttered molecular scenes? Discover label control in SAMSON.
When working on detailed molecular systems, clarity often makes the difference between insight and confusion. Labels—while useful—can sometimes clutter your 3D view, especially during presentation or when focusing on specific structures. Fortunately, SAMSON offers an elegant solution: precise control over…
Focusing on Function: How to Align Specific Protein Regions for Better Structural Insight
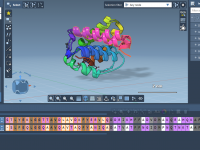
Speed Up Molecular Modeling with Attribute-Based Backbone Filters
When building or analyzing complex molecular systems, molecular modelers often face a recurring challenge: identifying specific fragments such as backbones with particular properties—like charge, composition, visibility, and more—across vast datasets or large molecular assemblies. Manually browsing through molecular groups wastes…