Making Atoms Appear When You Want Them To
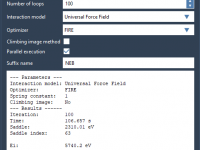
A Faster Way to Refine Molecular Transition Paths in SAMSON
Making Light Nodes Work For You in Molecular Design Projects
Quickly Identify and Select Visual Styles in SAMSON Using the Node Specification Language
When working on complex molecular simulations, efficient rendering management can make a significant difference in understanding and presentation. For modelers using SAMSON, keeping track of visual styles applied to different molecular models—like surfaces, ribbons, or atomistic representations—becomes more practical with…
Cleaning Up Protein Structures for Smooth Transitions
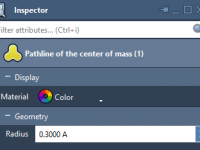
A Closer Look: How to Follow Atoms in Motion in SAMSON
Quickly Filter Molecular Animations with NSL Attributes in SAMSON
When working on molecular animations, molecular modelers often need to selectively visualize or analyze specific animation nodes based on their properties. However, navigating large molecular systems with multiple overlapping animations can get time-consuming and error-prone. This is where the Node…