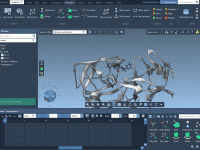
Creating smooth molecular presentations with SAMSON’s Animator panel
Sharing scientific discoveries effectively often means going beyond static visuals. Molecular modelers today are increasingly asked to create engaging and clear presentations, whether for publications, teaching, or collaboration. One recurring challenge is: how to animate molecular scenes smoothly, without spending…
Will SAMSON Run on My Operating System? Here’s What You Need to Know
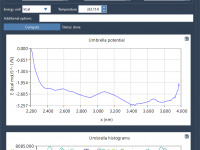
What Histogram Shapes Reveal in PMF Analysis
Faster Nanostructure Design with Pattern Editors in SAMSON
Your Scientific Profile is More Than a Username
Creating Molecular Docking Animations Without Code
The Quiet Timesaver: Interactive Tutorials in SAMSON
Why Did My Label Disappear? Understanding Label Visibility in SAMSON
Vertical Camera Moves Without the Guesswork: Mastering Pedestal Animations in SAMSON
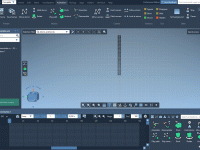
Creating smooth animations of complex molecular systems can be both powerful and time-consuming. One common challenge molecular modelers face is how to create clean, precise vertical camera motions—especially when communicating structural changes or navigating vertically extended biomolecular assemblies. Whether exploring…