Installing and Editing Local Python Packages Inside SAMSON
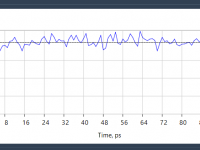
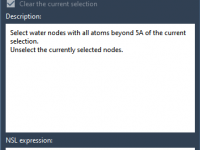
Many molecular modelers and computational chemists have their own Python packages—for custom simulations, analysis pipelines, or even neural networks for property prediction. But using them often means jumping between development environments, terminal sessions, and structured molecule viewers. What if everything…