Thinking of Developing a SAMSON Extension? Here’s What You Need to Know
Bring Depth to Your Molecule: Ambient Occlusion in SAMSON
Maintaining Focus While Moving Vertically in Molecular Animations
Selecting Initial Conformations for Batch Simulations in SAMSON’s GROMACS Wizard
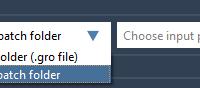
When launching molecular simulations, especially for systems with flexible conformations, many researchers face a common bottleneck: how to prepare multiple initial states for batch simulations efficiently without duplication of effort. Whether the goal is to explore conformational diversity or sample…
Collaborate Effortlessly: How Group Management Simplifies Molecular Modeling Workflows
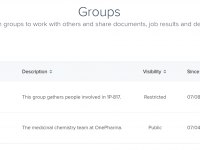
Working on molecular modeling projects often involves more than just simulations and structures — it’s also about sharing, collaborating, and managing contributions across institutions and teams. Scientists, educators, and developers frequently hit roadblocks when trying to organize access to shared…
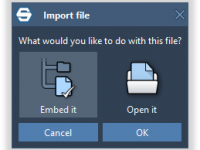
Making research files portable: embedding everything inside your molecular document
Animating Molecular Trajectories with SAMSON’s ‘Play Path’ Feature
Smoothly Animate Molecular Trajectories Between Keyframes in SAMSON
When working with molecular simulations, visualizing motion is essential to understand molecular mechanisms, reaction pathways, or conformational changes. Molecular modelers often run simulations that yield extensive trajectories—collections of molecular conformations over time. But how do you convert raw trajectory data…