When working with Protein Data Bank (PDB) files, molecular modelers often face a limitation: the structures frequently describe only asymmetric units—just a part of the full biological or crystal assembly. This can make it hard to visualize or simulate the full ensemble of protein-protein interactions, especially when working with quaternary structures or oligomers.
This is where the Symmetry Mate Editor in SAMSON can help. If you’ve ever wanted to reconstruct a full biological assembly from a PDB file, understand crystal packing, or explore symmetric interfaces, this tool provides a visual and interactive way to generate replicas based on symmetry records.
Preview and Generate Replicas Interactively
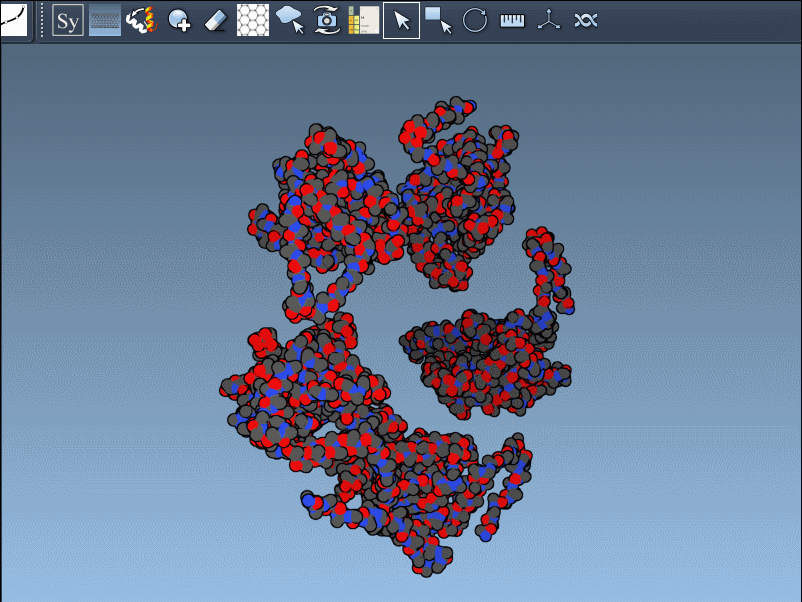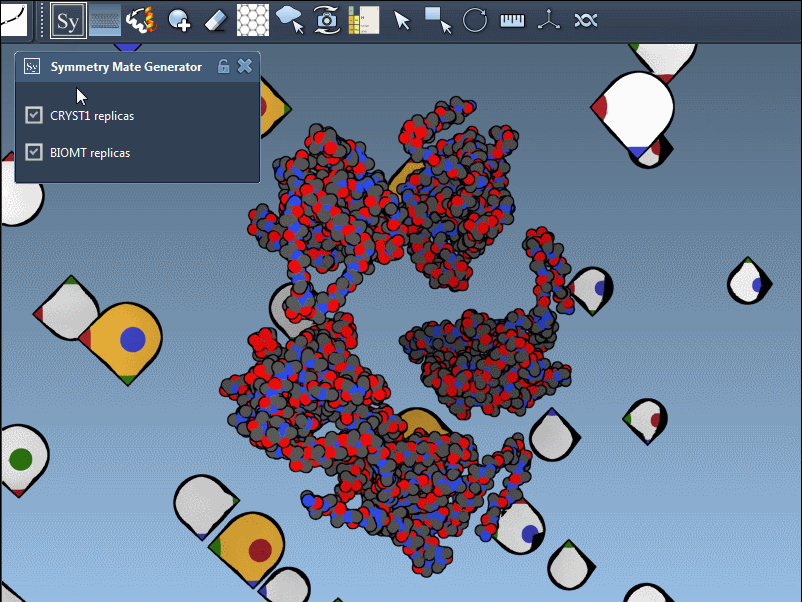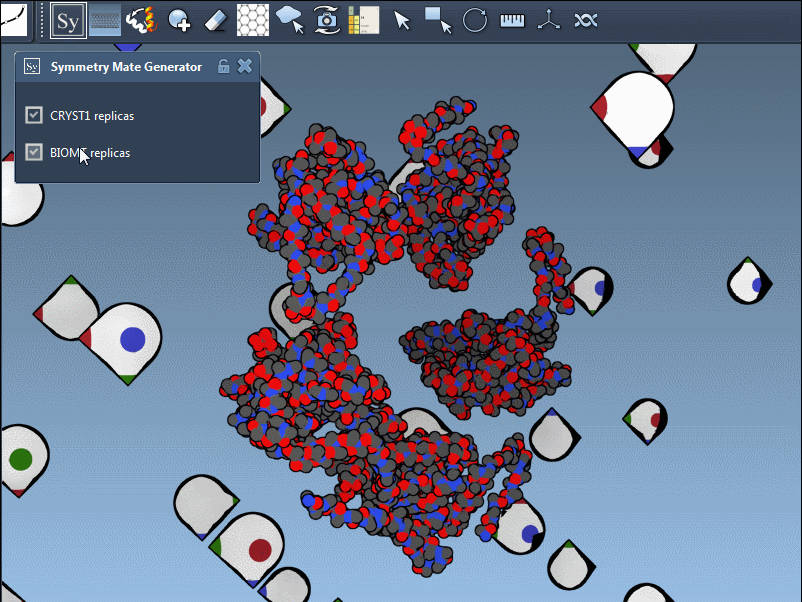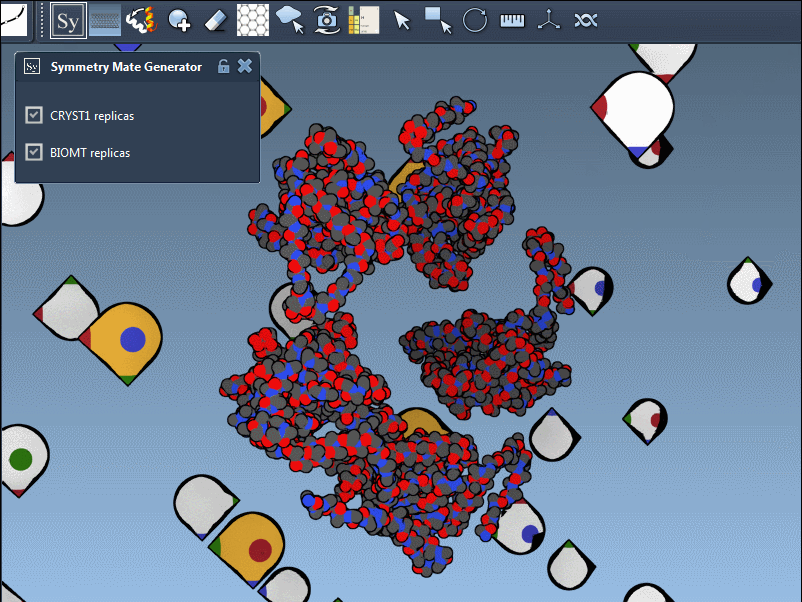
After activating the Symmetry Mate Editor from the menu or by pressing Shift + E, you’ll see control nodes in the 3D viewport. These represent symmetry transformations derived from CRYST1 (crystal packing) and BIOMT (biological assembly) records in the PDB file.
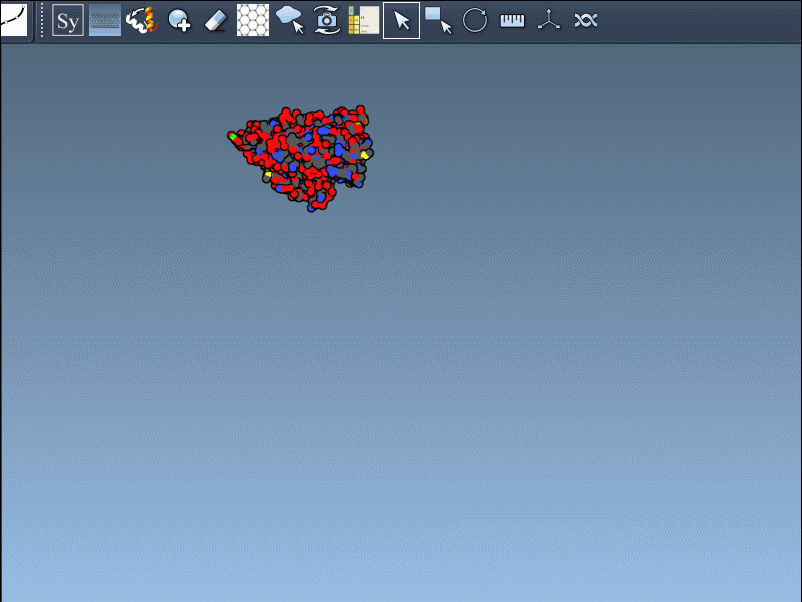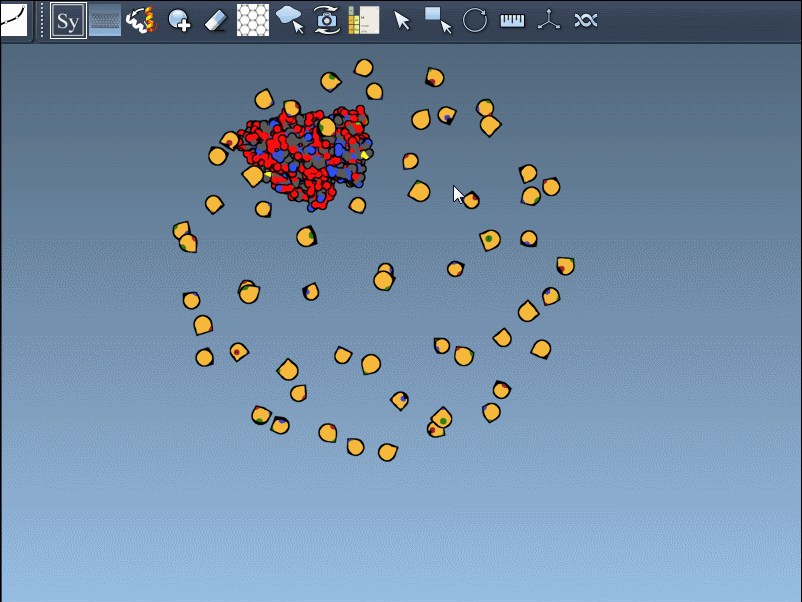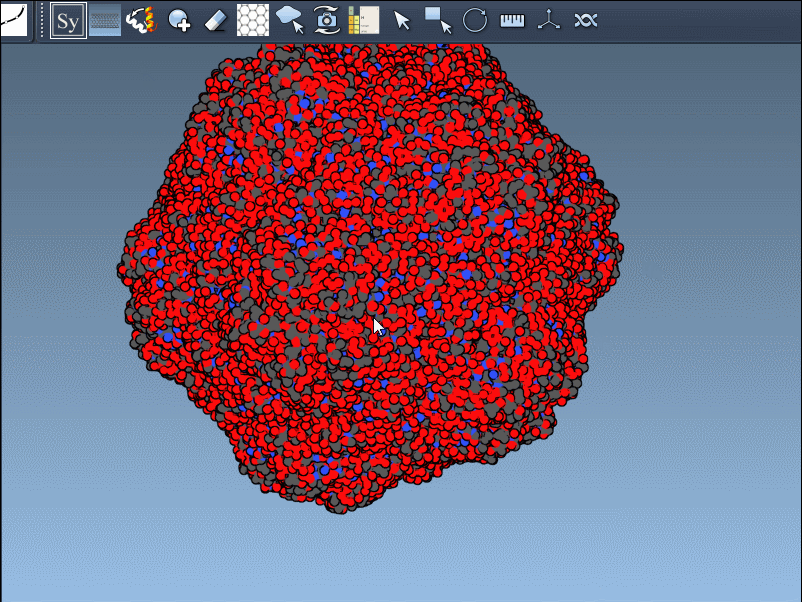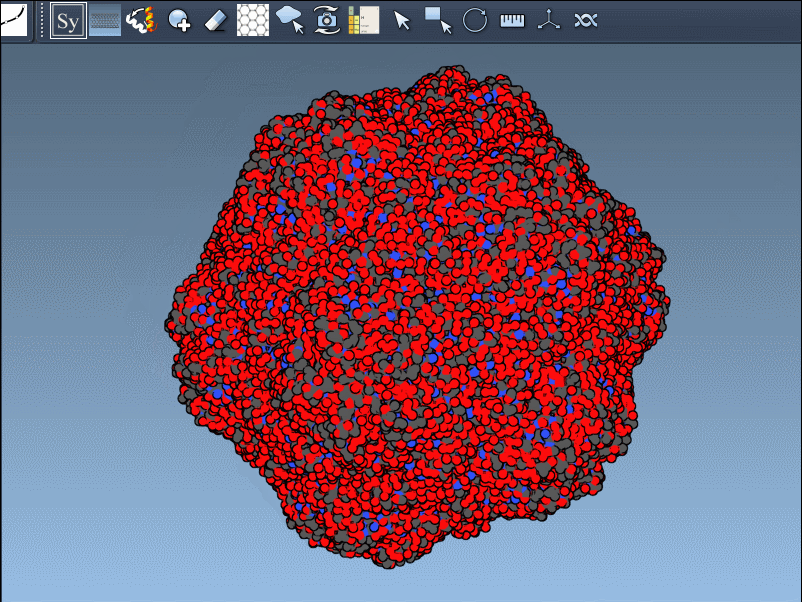
You can use Ctrl/Cmd + mouse wheel to explore and scale the number of control nodes. Hovering over a node gives you a real-time preview of the potential replica it will generate:
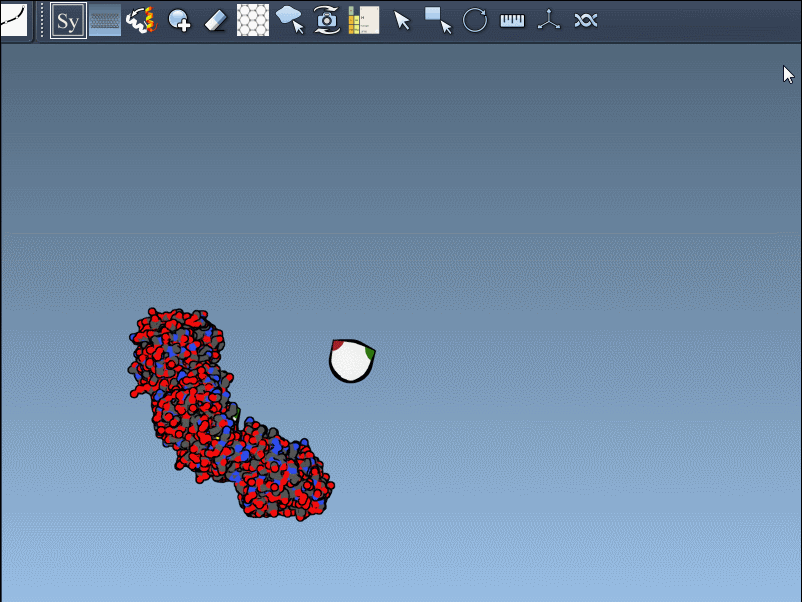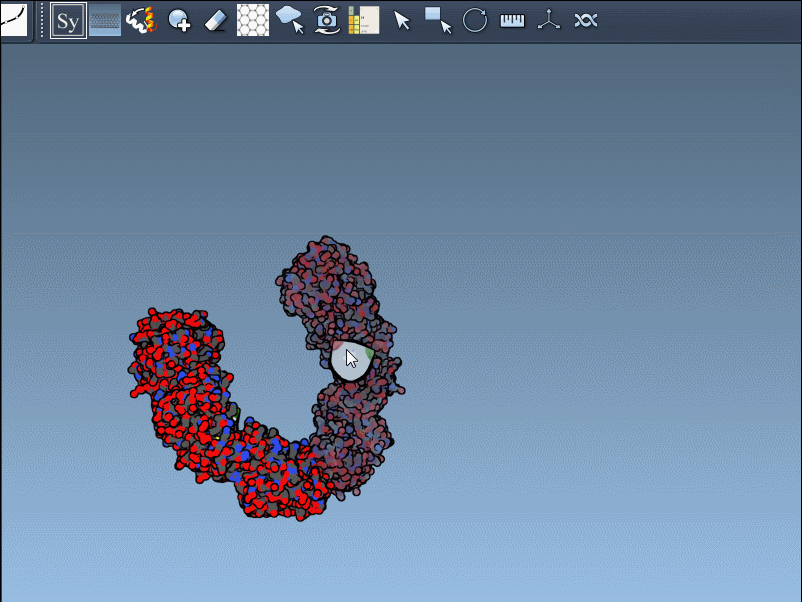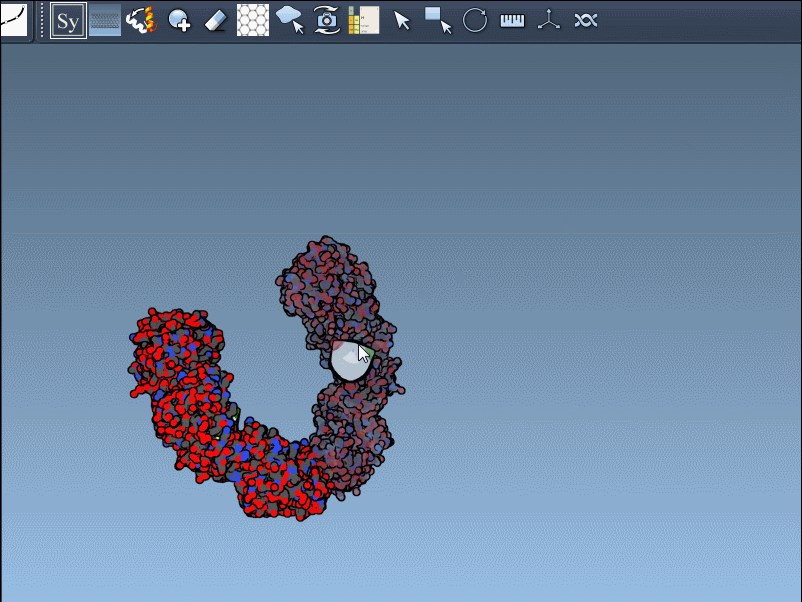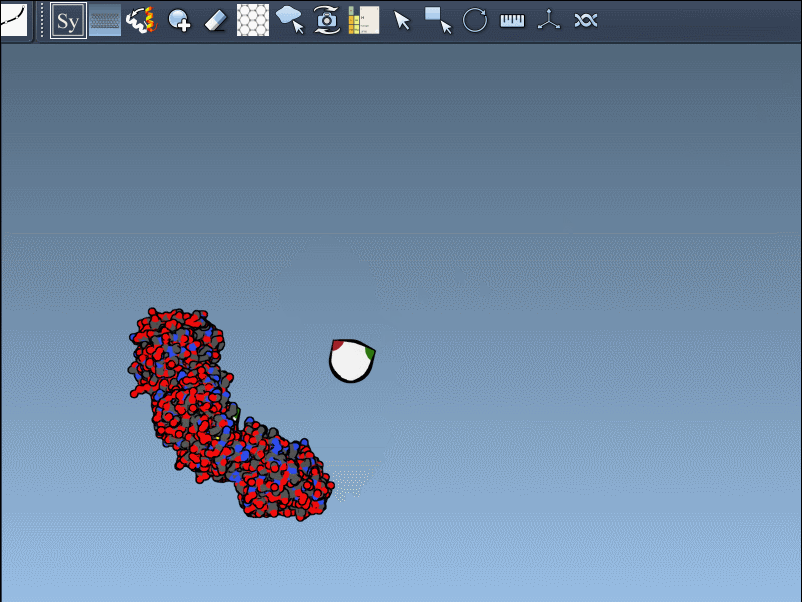
Click a node to generate that symmetry mate. Want to generate the full assembly in one go? Hold Ctrl/Cmd before clicking to activate all possible replicas at once:
CRYST1 vs. BIOMT: What’s the Difference?
The Symmetry Mate Editor distinguishes between two symmetry types, using distinct colors for clarity:
- CRYST1: Derived from the crystal lattice (white nodes)
- BIOMT: Based on biological assembly annotations (yellow nodes)
By toggling between these, users can visualize how a molecule is packed in the crystal vs. how it’s meant to function biologically. For example, the image below shows both types of assemblies for the 1B5S structure:
Typical Use Cases for Symmetric Replicas
- Visualizing full biological assemblies from asymmetric units
- Rebuilding symmetric oligomeric proteins for molecular dynamics simulations
- Designing symmetrical nanostructures or protein cages
- Identifying repeated functional sites across the structure
These reconstruction tasks are particularly valuable in docking studies or when preparing systems for long-timescale simulations.
Simple Controls, Clean Undo
Every replica you generate can be easily undone with Ctrl/Cmd + Z. Additionally, combining this tool with other extensions—like symmetry detection or ribbon rendering—can help extract more insight from the full assembly.
To learn more, visit the complete documentation page:
https://documentation.samson-connect.net/tutorials/symmetry/generating-symmetry-mates/
SAMSON and all SAMSON Extensions are free for non-commercial use. You can download SAMSON at https://www.samson-connect.net.





