If you’re running molecular dynamics simulations or exploring binding/unbinding pathways, you’ve probably encountered this question: where does the system actually go? While we often track individual atoms or align structures, it’s not always easy to get an intuitive grasp of how a group of atoms—or even a whole ligand—moves over time.
That’s where the Pathlines visual model in SAMSON comes in. With just a few clicks, you can visualize the motion of the center of mass (COM) of any group of atoms along computed paths. It’s especially useful for visualizing things like ligand unbinding, protein domain shifts, or diffusion events.
What Is a Center-of-Mass Pathline?
The motion of an atom group’s center of mass summarizes its trajectory in a clean, readable form. Rather than showing the position of every atom over time (which can quickly become messy), the COM pathline shows a single, smooth curve that represents the average weighted position of the selected atoms as they move.
This is particularly helpful when comparing multiple simulations, visualizing collective motions, or explaining results to collaborators.
Example: Ligand Unbinding Trail
Let’s walk through a quick example using a sample system available in SAMSON: the Lactose permease structure (1PV7) with its ligand Thiodigalactosid (TDG) and ligand unbinding paths. These paths were generated with another SAMSON Extension, Ligand Path Finder.
Once loaded, here’s how to visualize the center-of-mass pathline of the ligand:
- Open the Document view and select the ligand (named
TDG) and a path. If you’re on Windows, hold Ctrl while clicking; on macOS, use Cmd. - Navigate to Visualization > Visual model > More… or use the shortcut Ctrl/Cmd + Shift + V.
- From the dialog, choose Pathline of the center of mass and confirm with OK.
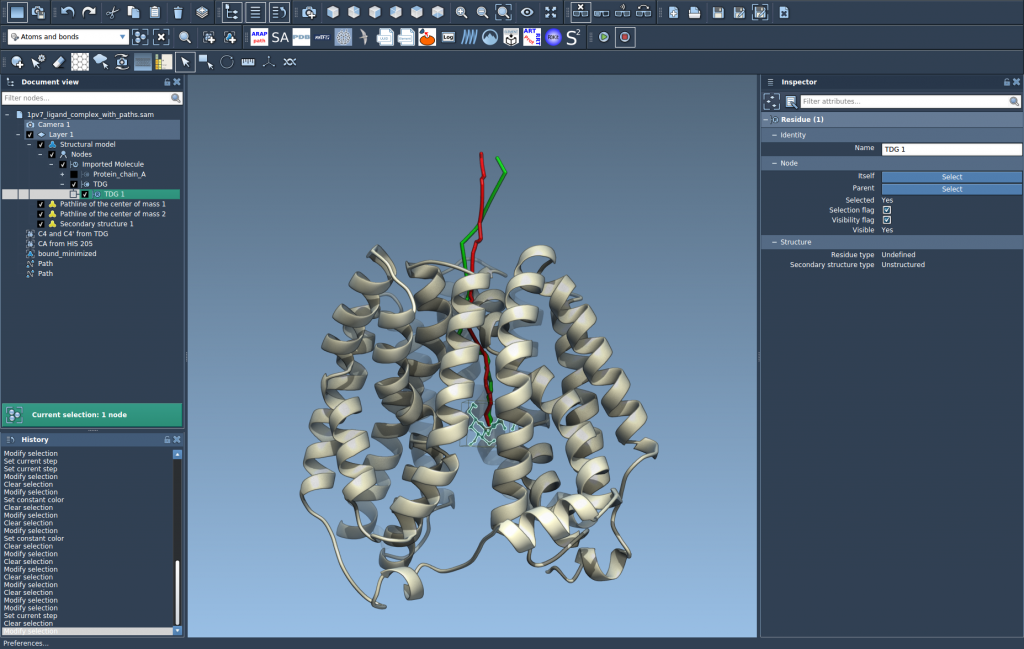
That’s it! You’ll immediately see a curved line that tells you how the ligand (or any selected group of atoms) moved along the chosen path.
Tips for Customizing the View
- You can adjust color and line thickness using the Inspector (Ctrl/Command + 2).
- Double-click a path to control animation, or right-click it for more options.
- If you’re running multiple paths, you can compare different COM trajectories at once.
Why This Matters
Many molecular modelers spend time trying to analyze and communicate how molecules move—but traditional RMSD plots or raw trajectories aren’t always intuitive. A center-of-mass pathline is a powerful visualization for:
- Comparing unbinding trajectories across conditions or ligands
- Showing collective domain motions in protein systems
- Quickly identifying irregular or unexpected paths
All it takes is a couple of selections and a couple clicks, so you can focus more on interpreting scientific meaning and less on technical setup.
To learn more and explore the full workflow, visit the complete Pathlines documentation.
SAMSON and all SAMSON Extensions are free for non-commercial use. You can get SAMSON at https://www.samson-connect.net.





