In molecular modeling, finding biologically relevant transition pathways between molecular states can be a challenge—especially when simulating ligand unbinding events. Often, you may already have a trajectory generated through sampling or interpolation, but need to make it more accurate from an energy perspective. This is where the Parallel Nudged Elastic Band (P-NEB) method in SAMSON becomes particularly useful.
This post will guide you through using P-NEB in SAMSON to improve ligand unbinding pathways by optimizing them into minimum energy paths. The method is applicable when you’re starting from a trajectory (called a path in SAMSON) and want to achieve a more physically meaningful and smooth transition between conformations.
Why optimize a path?
Transition path sampling or interpolation can give you rough trajectories, but they might not reflect the true energy landscape of the system. P-NEB helps by adjusting intermediate conformations to follow a minimum energy path, while maintaining proper spacing between them. This is done by introducing virtual spring forces that keep the images (conformations along the trajectory) evenly spaced and then optimizing using a force field like UFF.
Getting started
First, download a sample system from SAMSON’s Home > Download menu. Two examples are available:
- Zinc ligand unbinding trajectory (a smaller system for testing)
- Lactose permease (1PV7) with TDG ligand unbinding paths

These documents include pre-computed paths you can optimize further using P-NEB.
Launching and configuring P-NEB
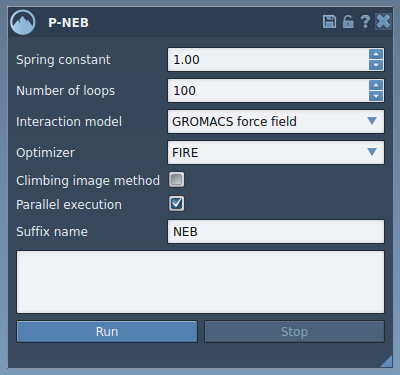
Open the P-NEB app via Home > Apps > All > P-NEB. In the interface, configure the following fields:
- Spring constant: 1.00
- Number of loops: 100
- Interaction model: Universal Force Field (UFF)
- Optimizer: FIRE
- Climbing image method: Leave unchecked (try later if needed)
- Parallel execution: Checked
- Suffix name: NEB
Applying P-NEB to your path
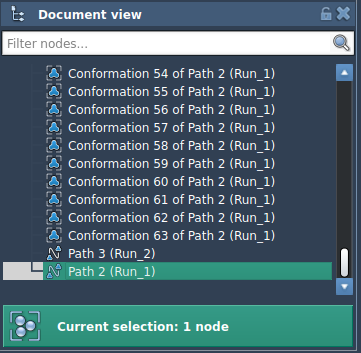
In the Document View, select the path node for the unbinding trajectory. Then, click Run in the P-NEB app. The app will ask whether to use existing bonds—confirm with “Yes”. The optimization process will begin, showing progress on the status bar.

Reviewing results
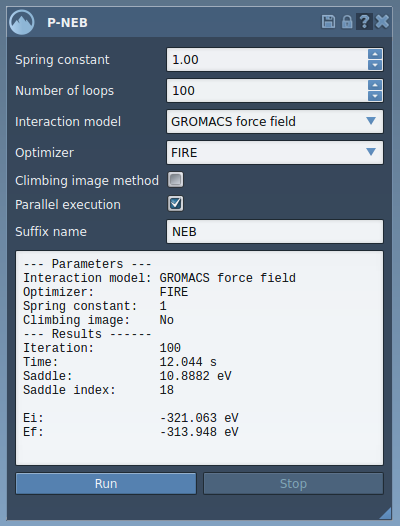
Once complete, a new path will appear in the Document View. You can double-click it to animate the optimized transition, or inspect the structural changes in more detail using the Inspector panel. The original path remains unchanged, so you can compare the trajectories side-by-side.
When to use this feature
If you have an existing unbinding, binding, or conformational pathway generated via sampling, machine learning, or other path-generation tools in SAMSON (e.g., Ligand Path Finder), P-NEB provides a way to refine those paths into more realistic trajectories.
This method is especially recommended when your goal is not just to visualize transitions, but to inspect likely energetic barriers or intermediates along the path—such as for estimating kinetics or designing new ligands.
Conclusion
Transforming rough transition data into detailed, energetically meaningful pathways can save countless simulation hours and help identify key interactions along binding or unbinding processes. P-NEB in SAMSON offers a flexible and user-friendly approach to tackle this challenge.
To learn more and go deeper into options such as climbing image mode or conformational optimization, visit the full documentation: Optimize transition paths with the Parallel Nudged Elastic Band method.
SAMSON and all SAMSON Extensions are free for non-commercial use. You can download SAMSON at https://www.samson-connect.net.





