If you’ve ever worked on optimizing a lead compound, you know how valuable small molecular changes can be. What if you could quickly and systematically generate analogs by swapping a single atom or group—right from your molecular design platform?
This post introduces a concise and visual workflow in SAMSON for generating analogs of your molecules using Positional Analogue Scanning via the SMILES Manager. The technique helps you explore structure-activity relationships (SAR) by examining the impact of localized substitutions—such as replacing an aromatic hydrogen with nitrogen or adding a methyl group. This may affect how your compound binds, its solubility, or even its selectivity, and it all starts with one molecule.
Start from One Molecule
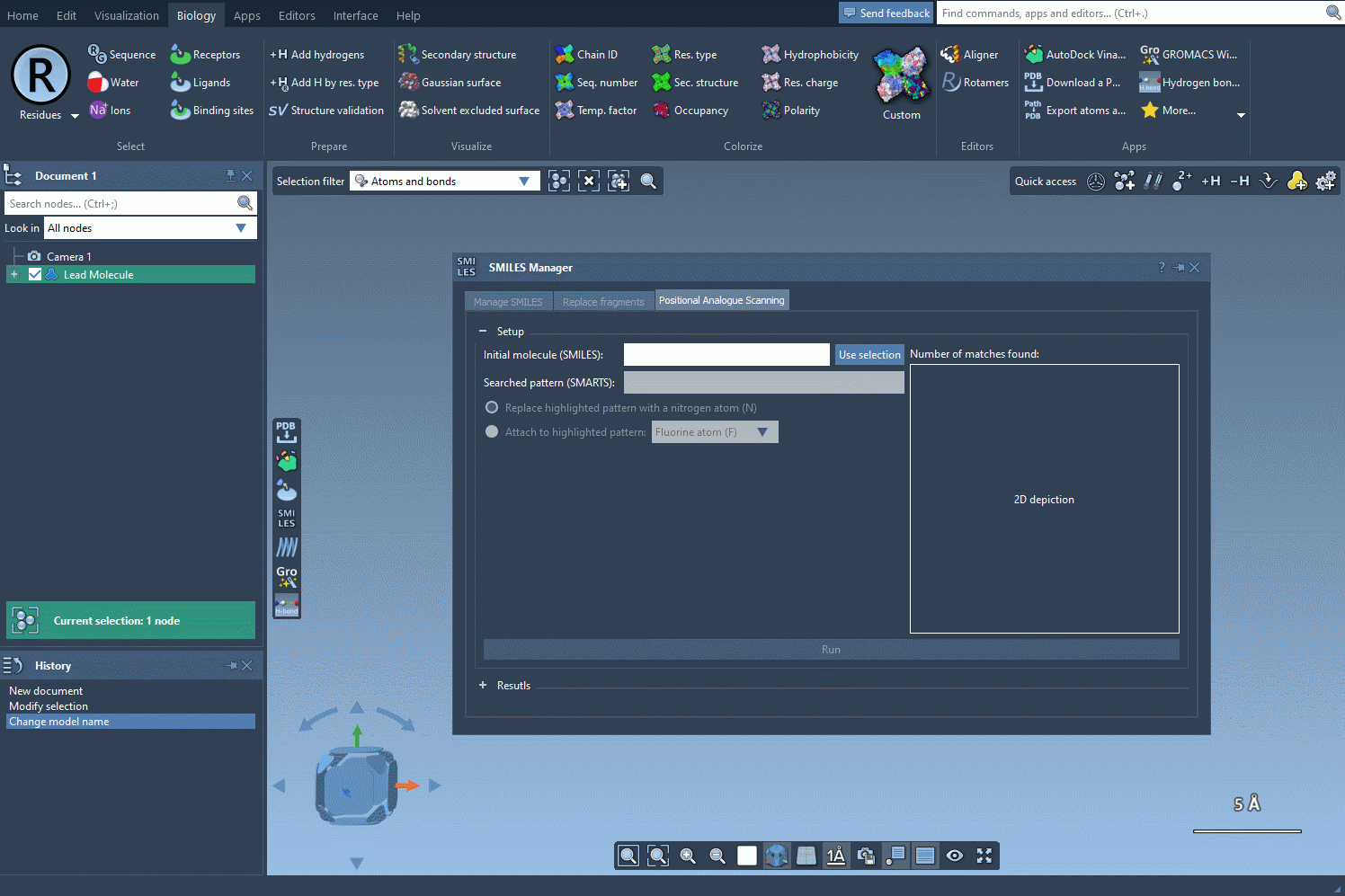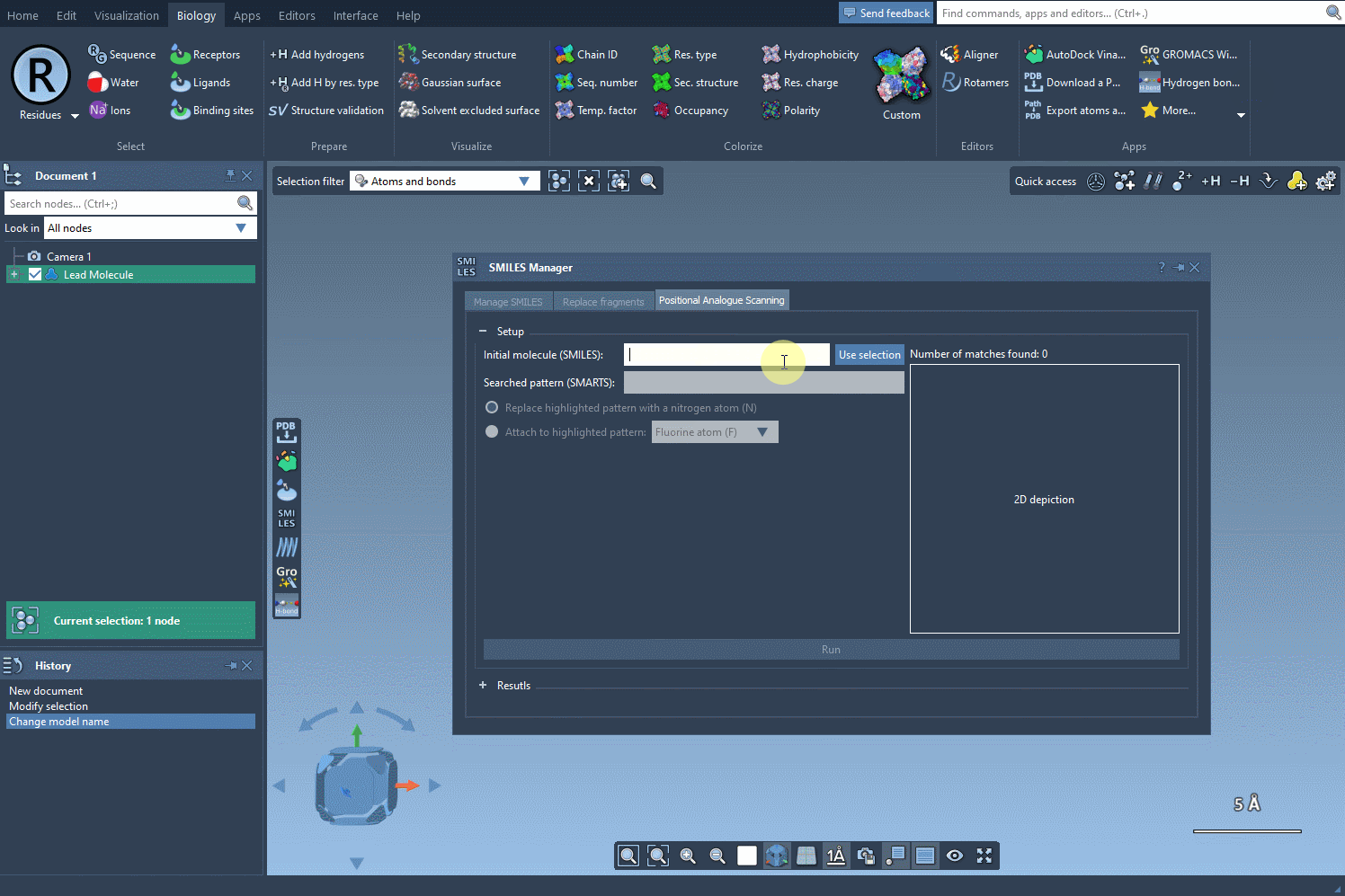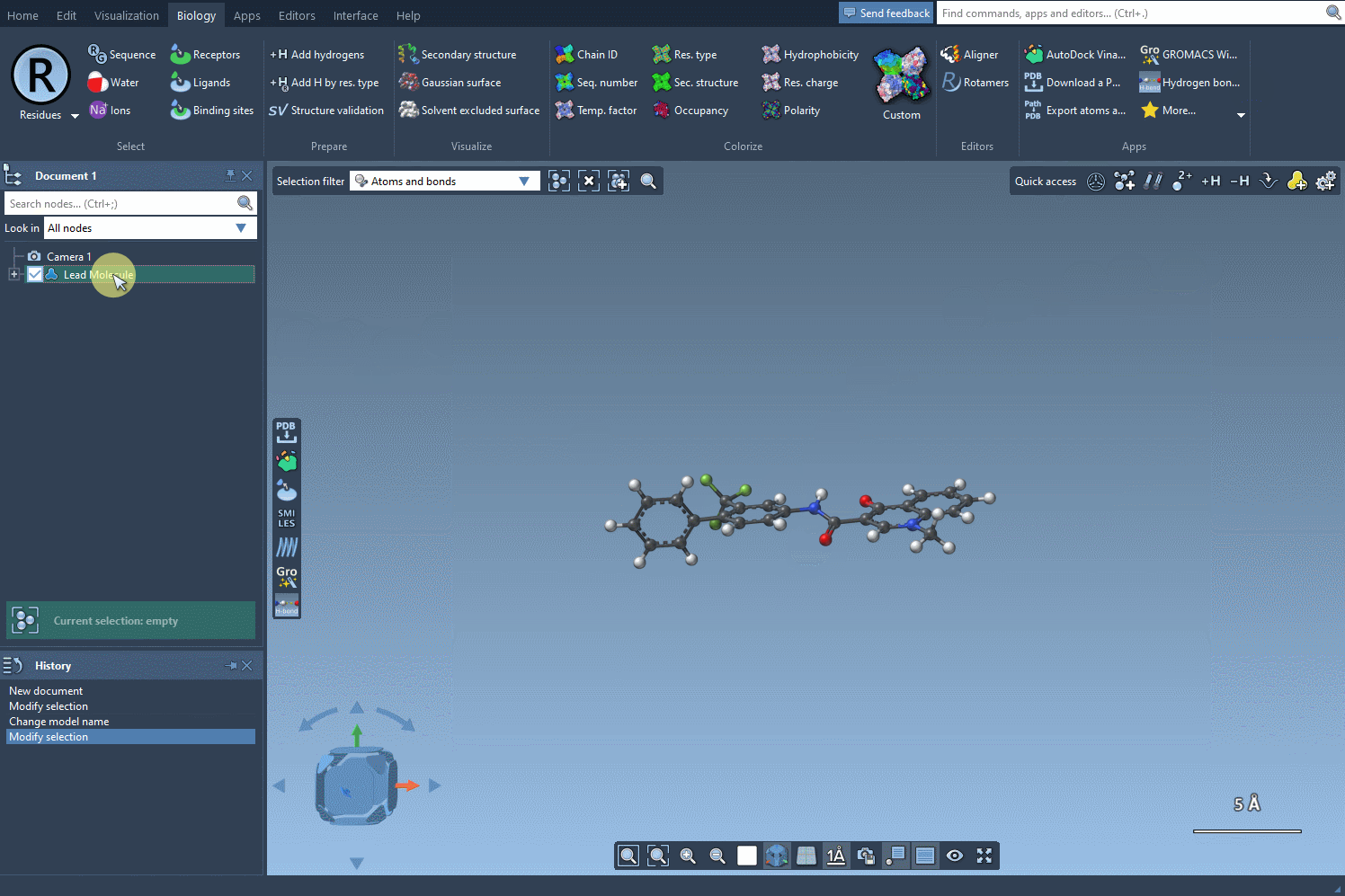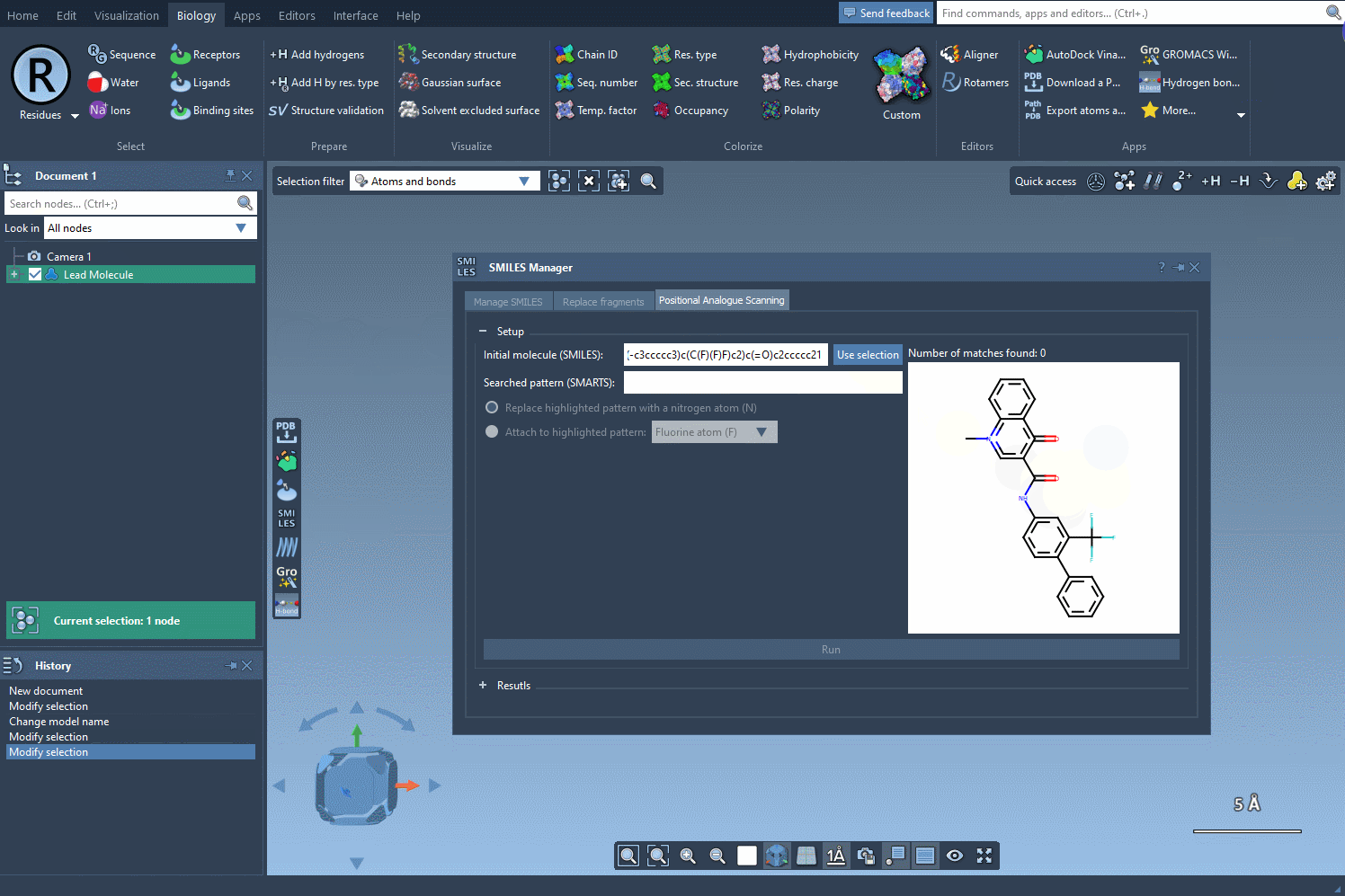
Begin with a molecule you’re looking to improve. In SAMSON, you can define your molecule by either pasting its SMILES code directly or selecting it from your document and clicking on the Use selection button. The interface is intuitive, letting you immediately move to defining patterns for substitution.
Define What to Modify
Next, specify which atoms or chemical features you want to scan. For instance, if you want to scan all aromatic hydrogens, just input the SMARTS pattern [cH]. SAMSON automatically highlights each match in your molecule, so you can confirm exactly where the changes will occur.

Attach or Replace
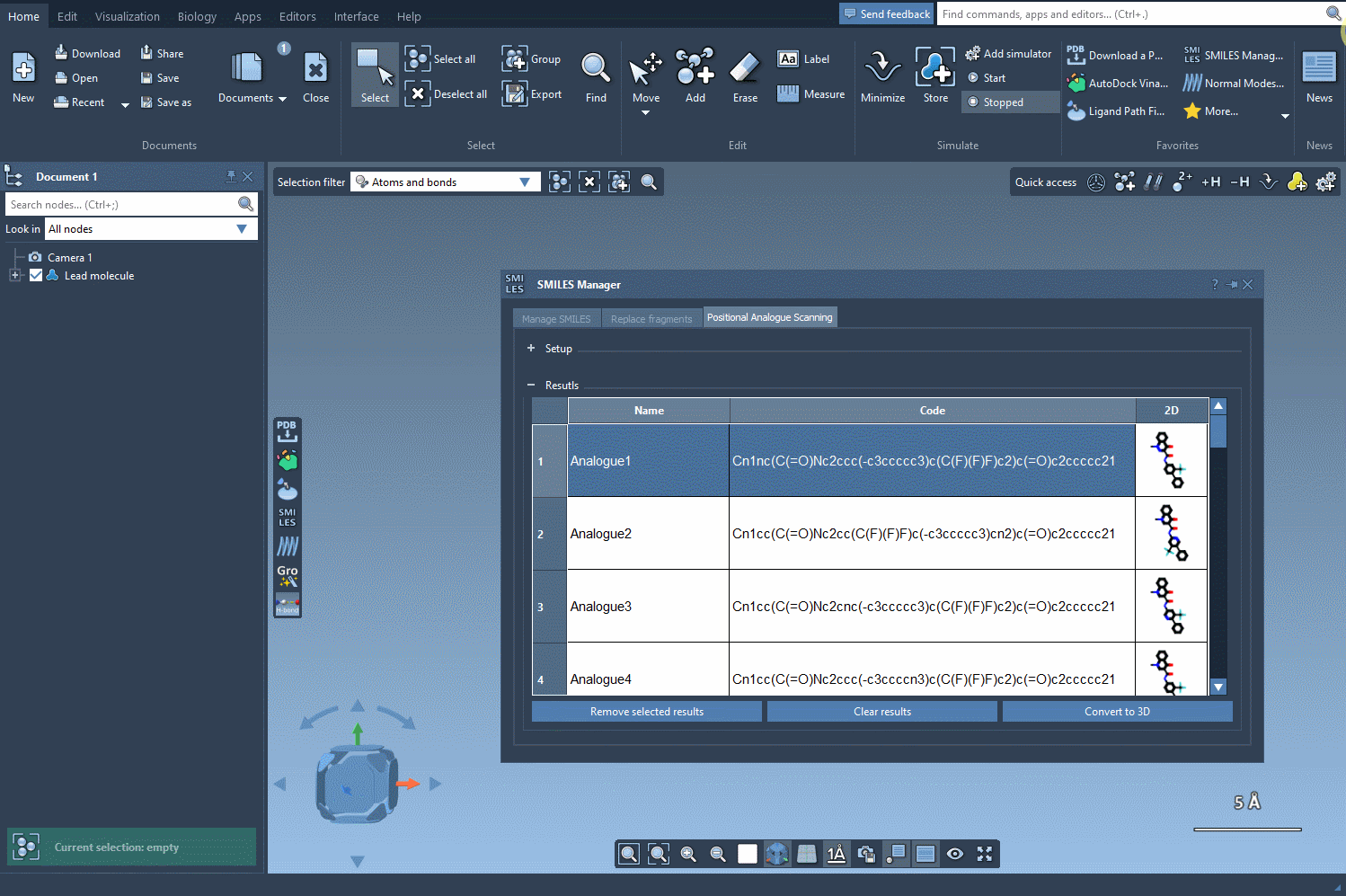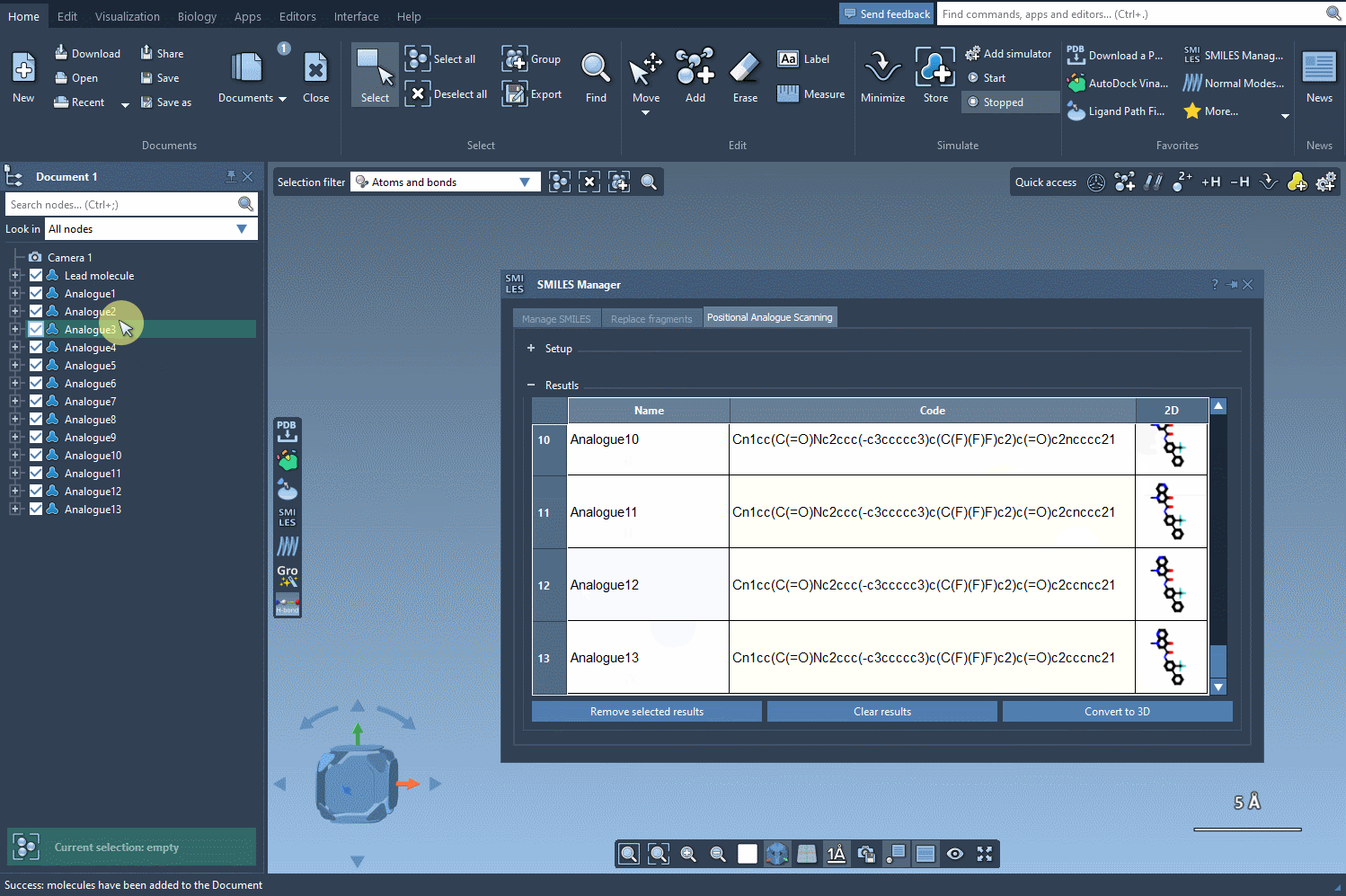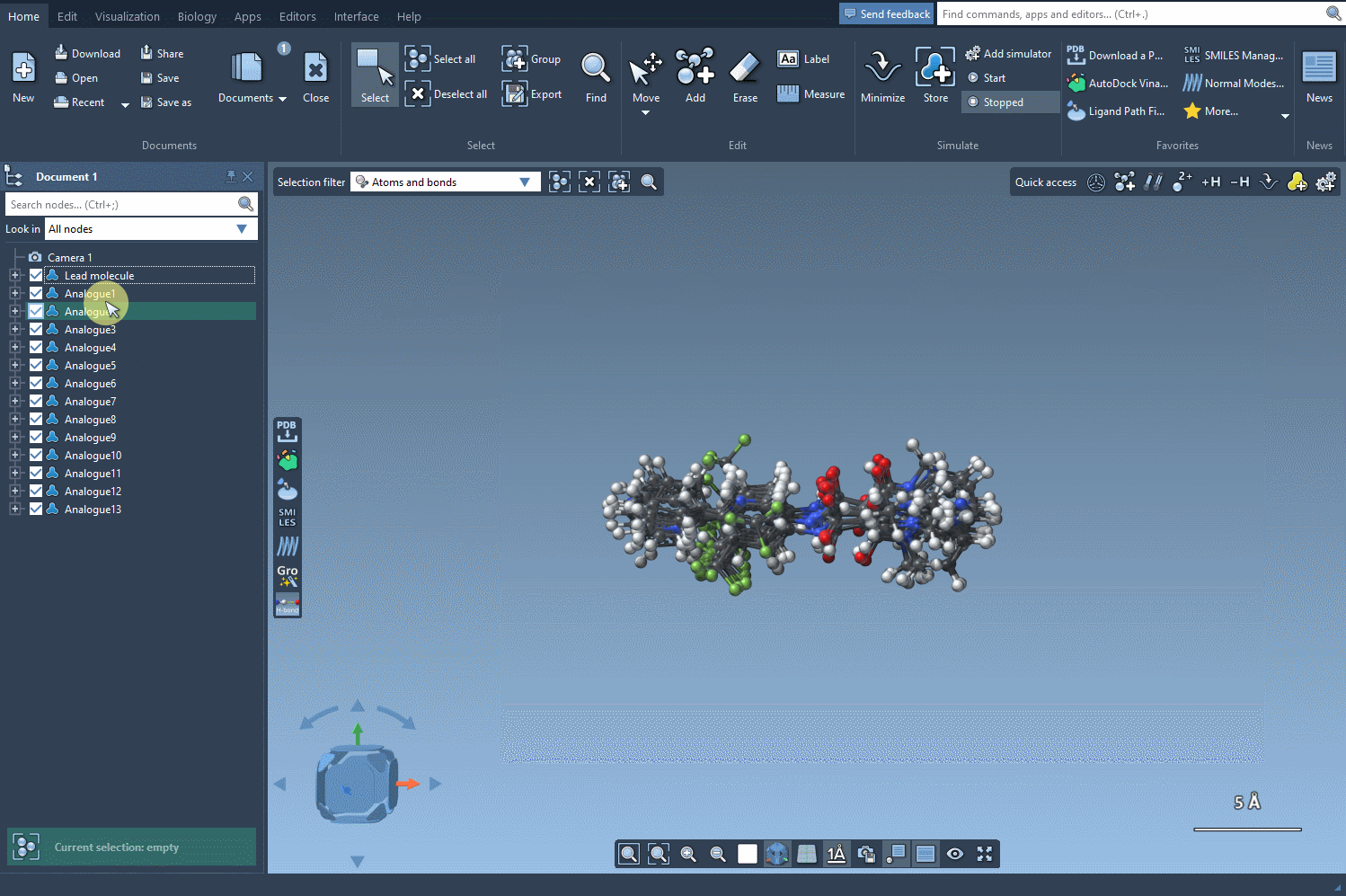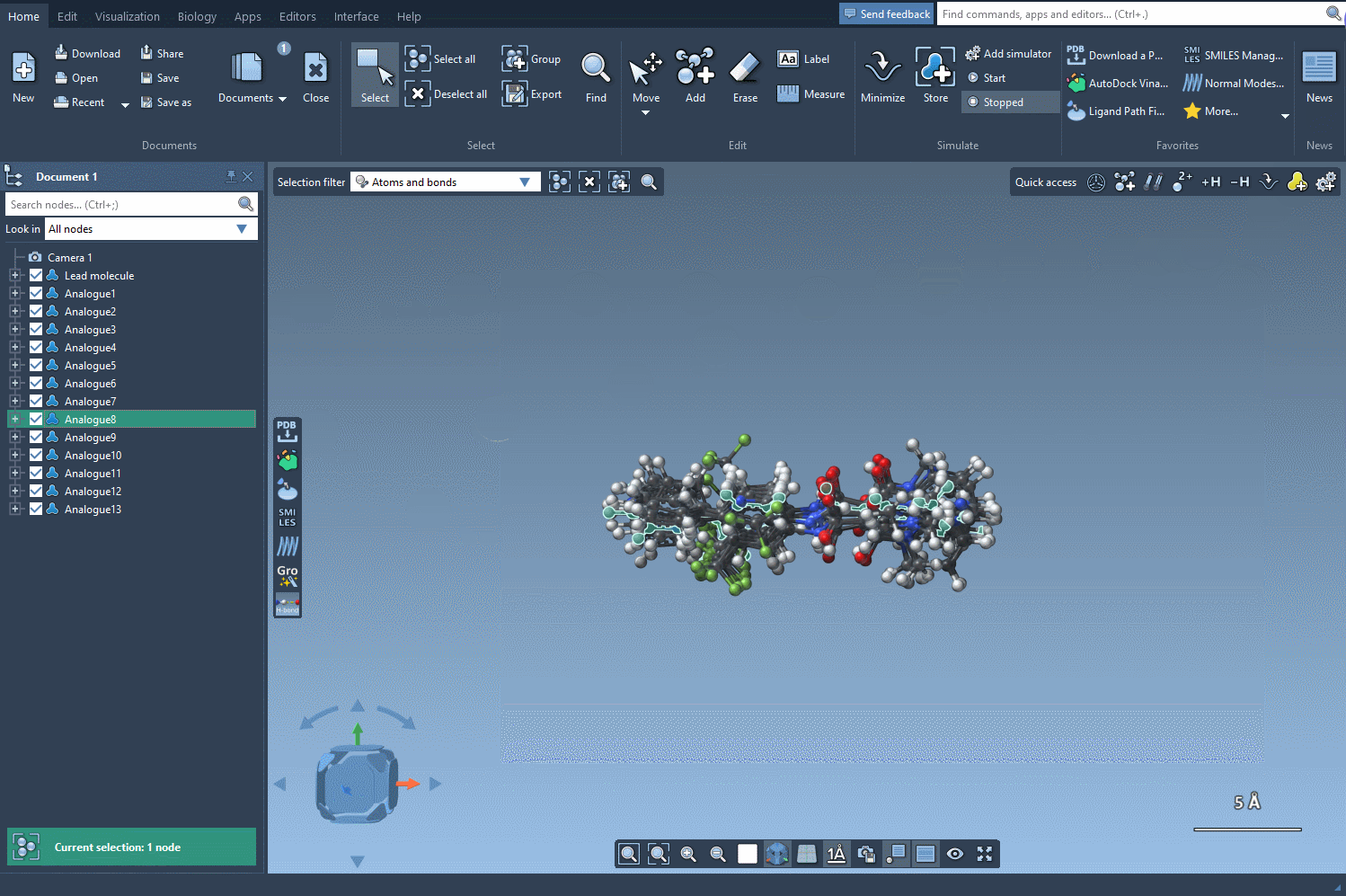
You now choose the modification: either replace the highlighted atoms or attach a new group to them. For example, swap aromatic hydrogens with nitrogen atoms, or attach a fluorine or methyl group. Click Run and your analogs will be automatically generated—with both 2D representations and their SMILES codes neatly displayed.
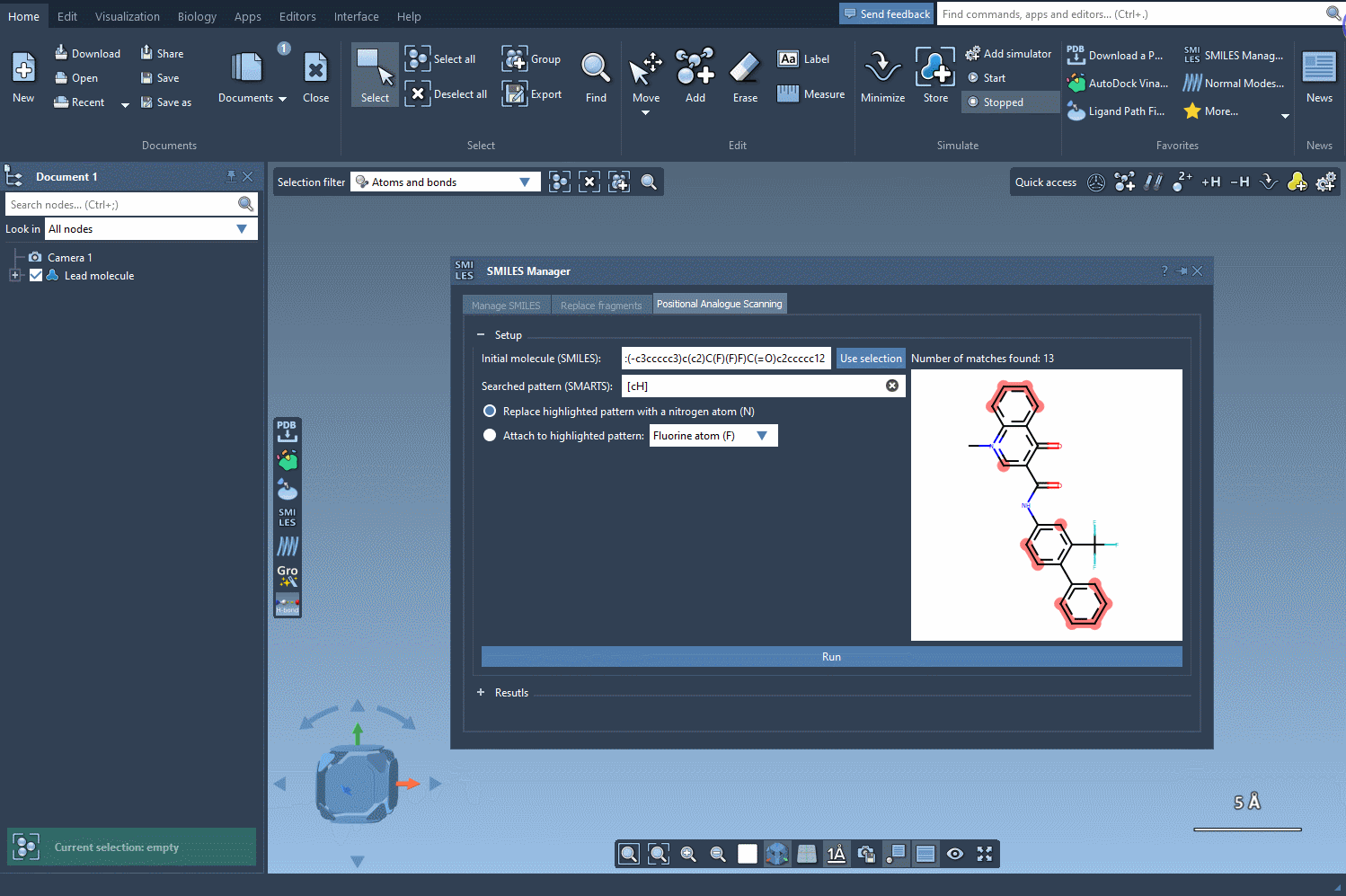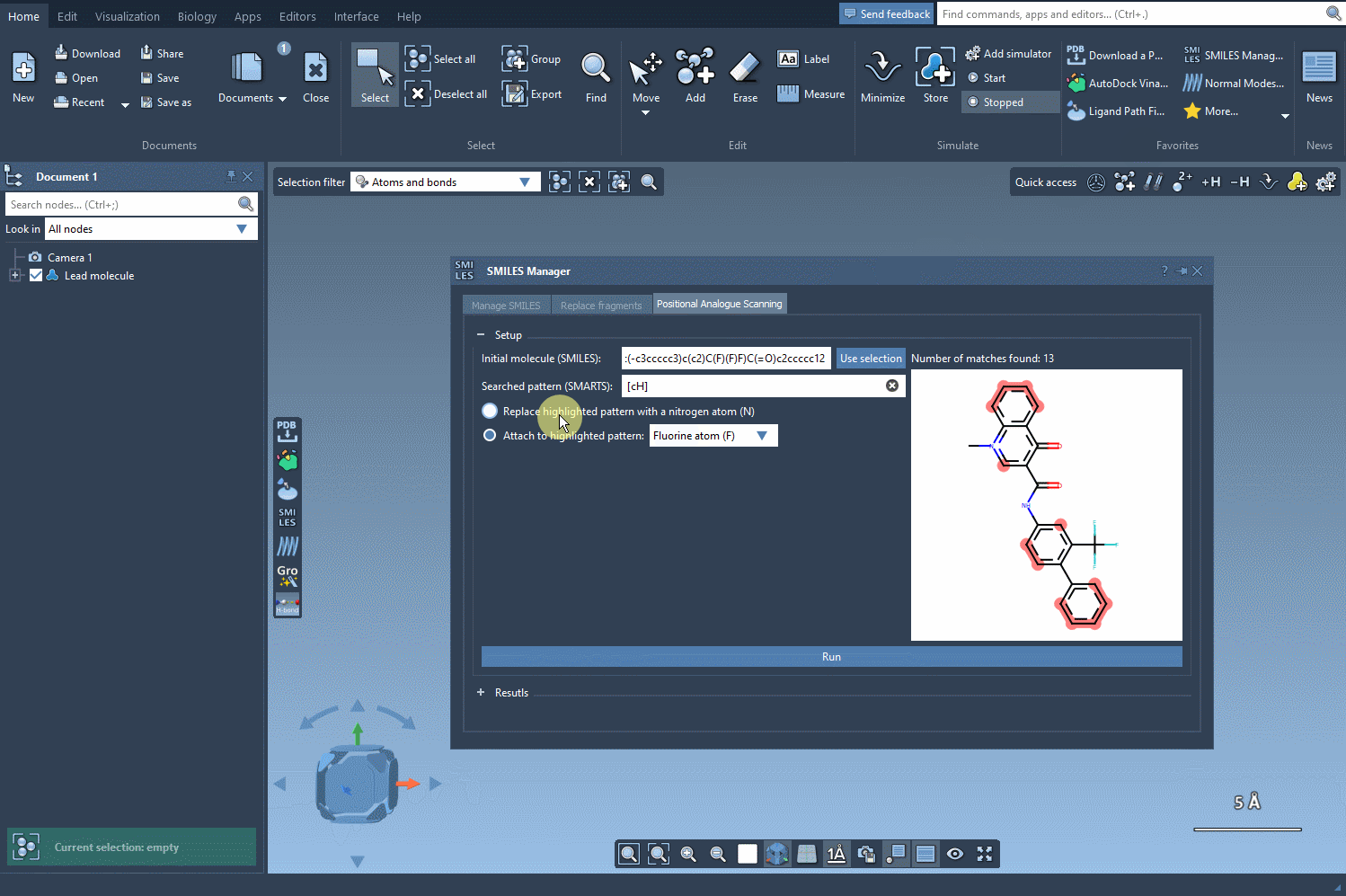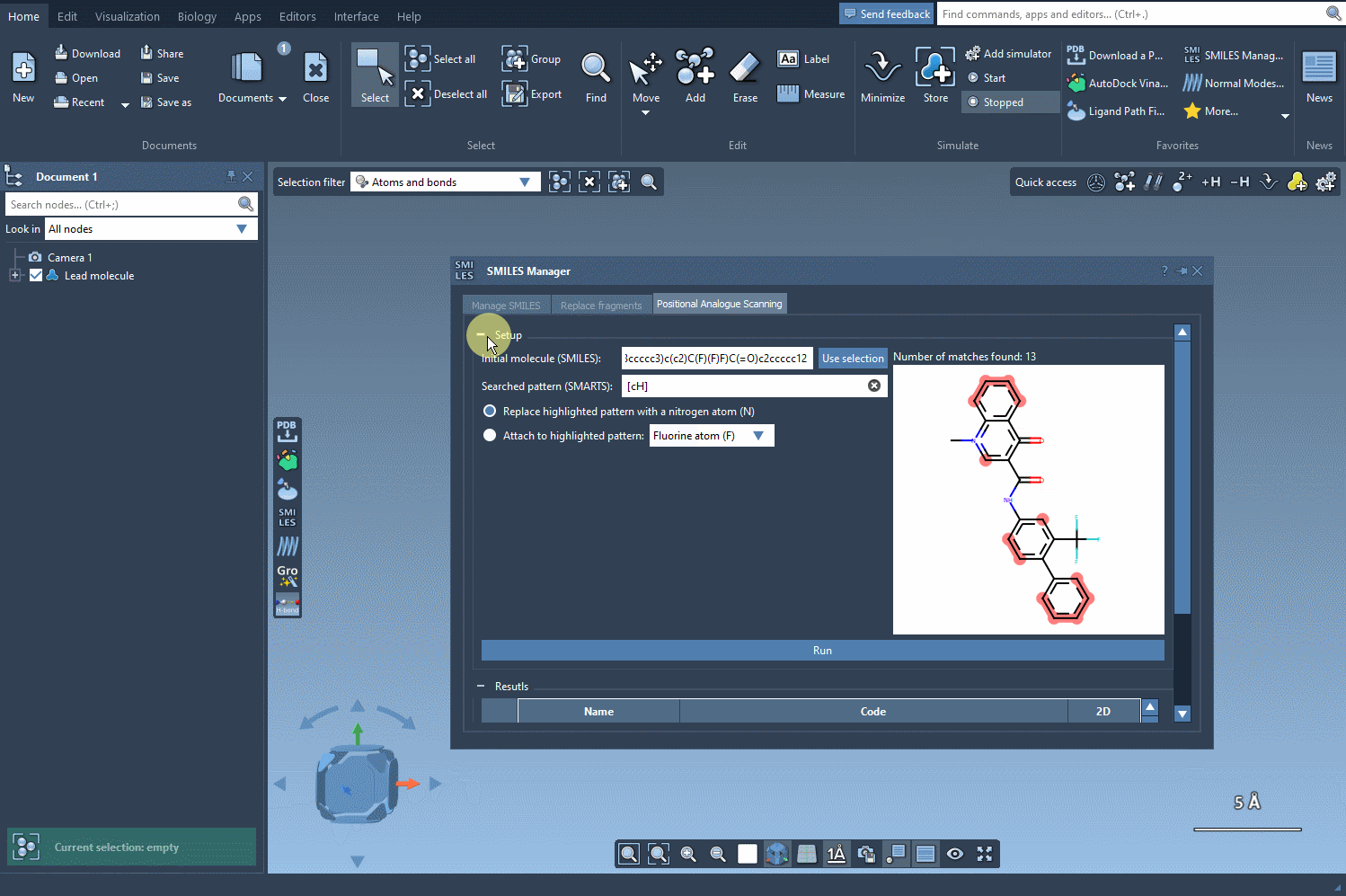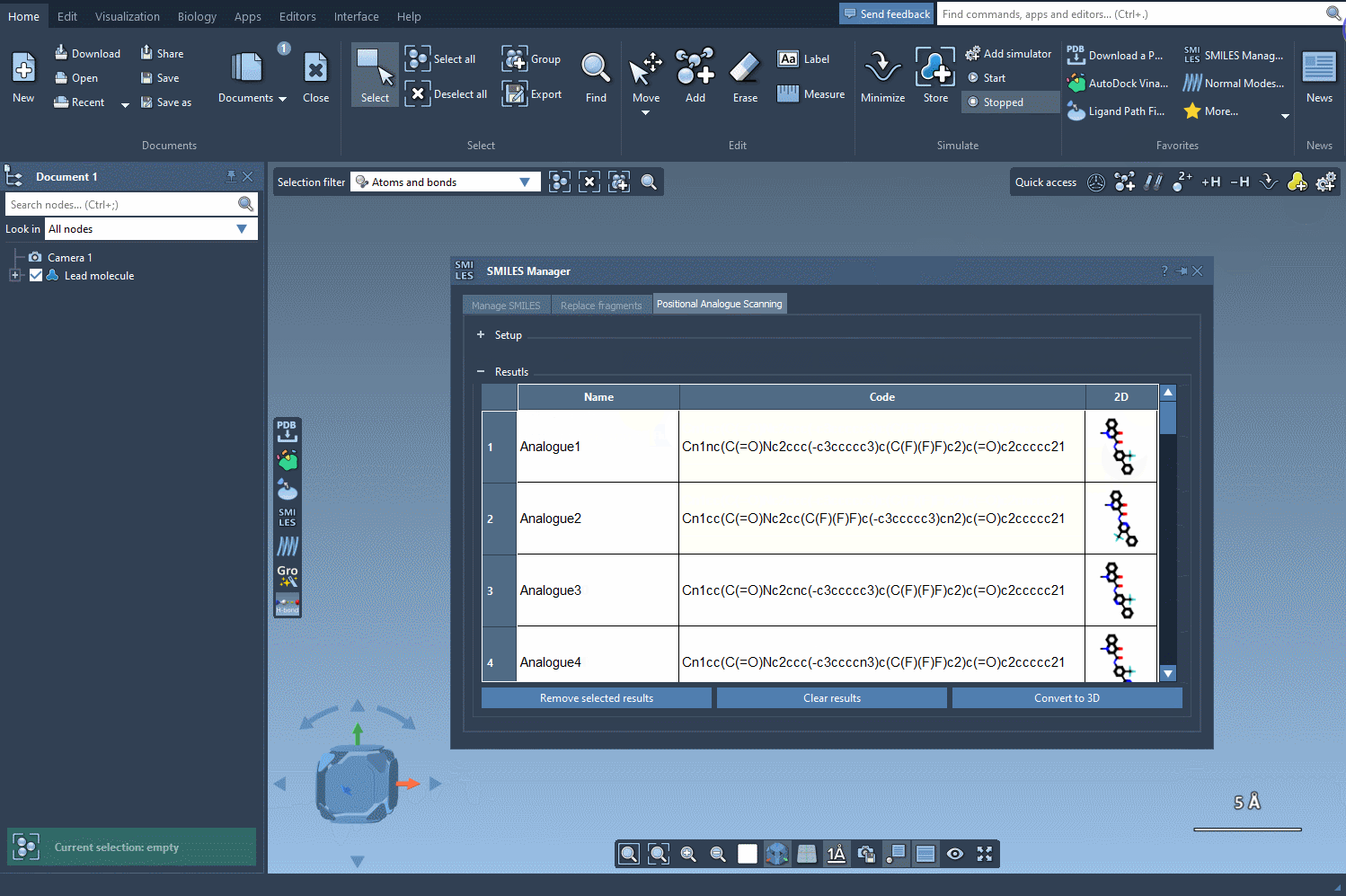
Explore and Edit Your Analog Table
The result table is more than just a list. You can:
- Rename or edit any analog by double-clicking.
- View larger 2D images with a double-click.
- Toggle image display for a clearer overview.
- Generate 3D structures for selected analogs with a simple right-click.
- Remove unwanted analogs individually or clear the table completely.
Next Step: 3D Structures and Further Investigation
Once you’ve generated analogs, click the Convert to 3D button to build their 3D structures directly in SAMSON. This enables immediate use in downstream tasks like docking simulations with the Autodock Vina Extended extension. You can examine how structural tweaks influence binding to your target protein—without ever leaving the SAMSON environment.
If you’re balancing molecular design, intuition, and time, this feature gives you room for all three. It helps you quickly test “what if” scenarios and focus on the most promising changes for your next synthesis or simulation run.
To discover the full tutorial and more examples, visit the official documentation page: Perform Positional Analogue Scanning using the SMILES Manager in SAMSON.
SAMSON and all SAMSON Extensions are free for non-commercial use. You can download SAMSON at https://www.samson-connect.net.





