One of the frustrating challenges in molecular modeling is understanding and simulating large conformational changes in biomolecular complexes. For researchers working on viral mechanisms or drug discovery, visualizing the conformational transition of key viral components—such as the spike glycoprotein on SARS-CoV-2—can dramatically impact both communication and research planning.
The SARS-CoV-2 spike protein is the linchpin of viral entry into human cells. Its ability to switch between a closed (non-binding) state and an open (binding) state allows it to interact with the human ACE2 receptor, triggering infection. Capturing this dynamic motion isn’t straightforward: high-resolution static structures exist, but seeing the transition between these states typically requires tedious manual manipulation or substantial simulation resources.
This is where the SAMSON platform provides a helpful shortcut. Using two experimentally determined spike structures—6VXX (closed state) and 6VYB (open state)—the SAMSON team computed an interpolated motion that enables users to visualize the spike’s conformational transition. For molecular modelers and educators, this is particularly valuable. You can now show the movement of the spike in relation to receptor-binding events, enhance your presentations, or build more realistic docking scenarios.
The animations are available from three key viewpoints:
- Side view:
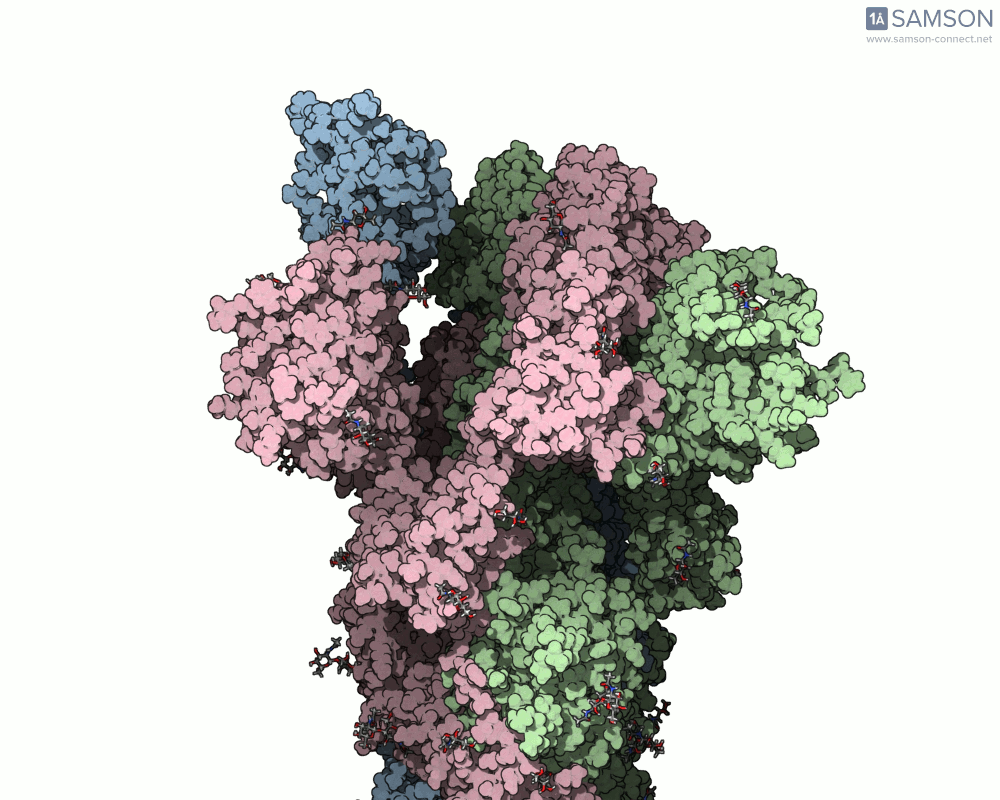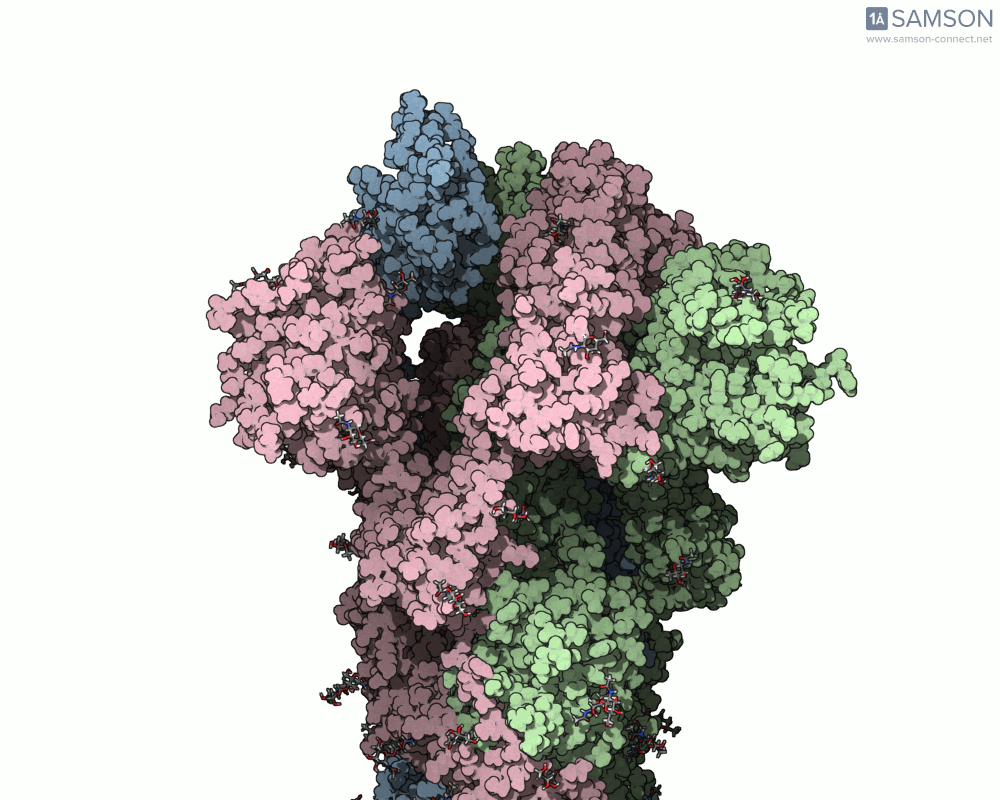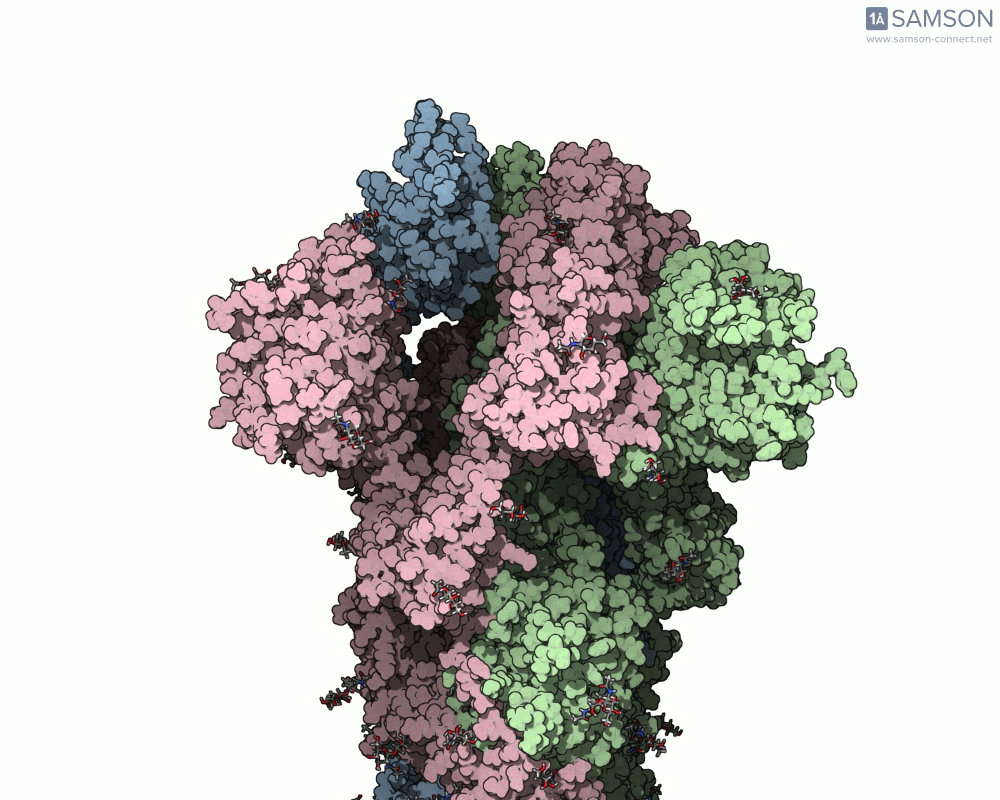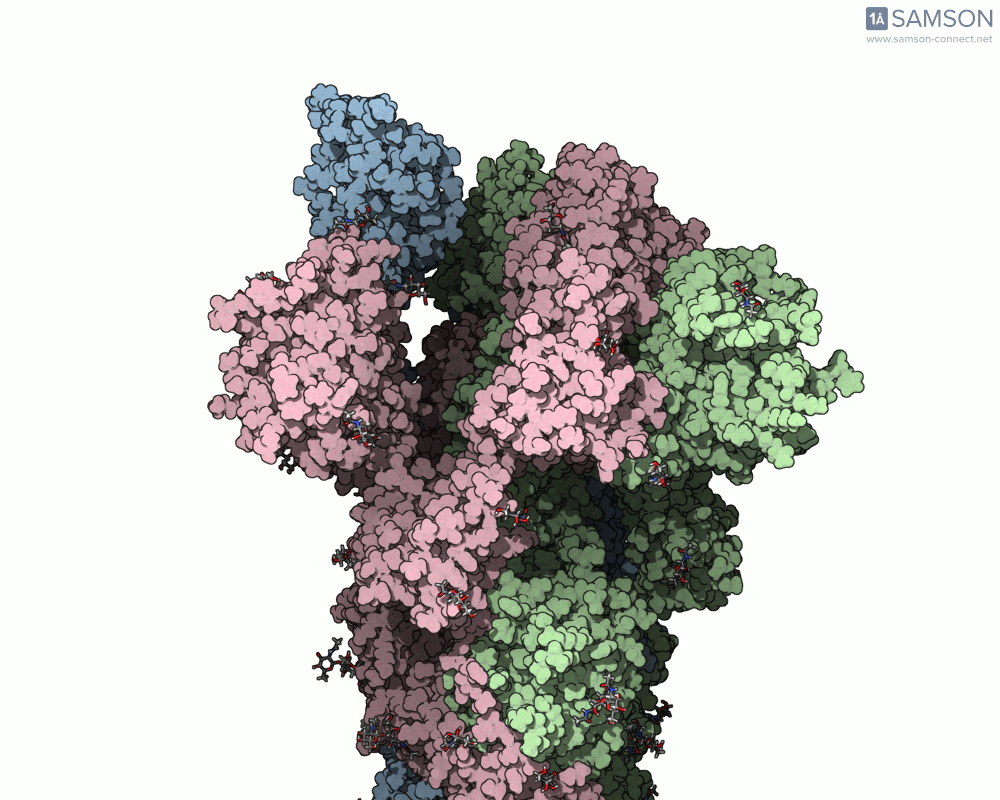
- Angular view:
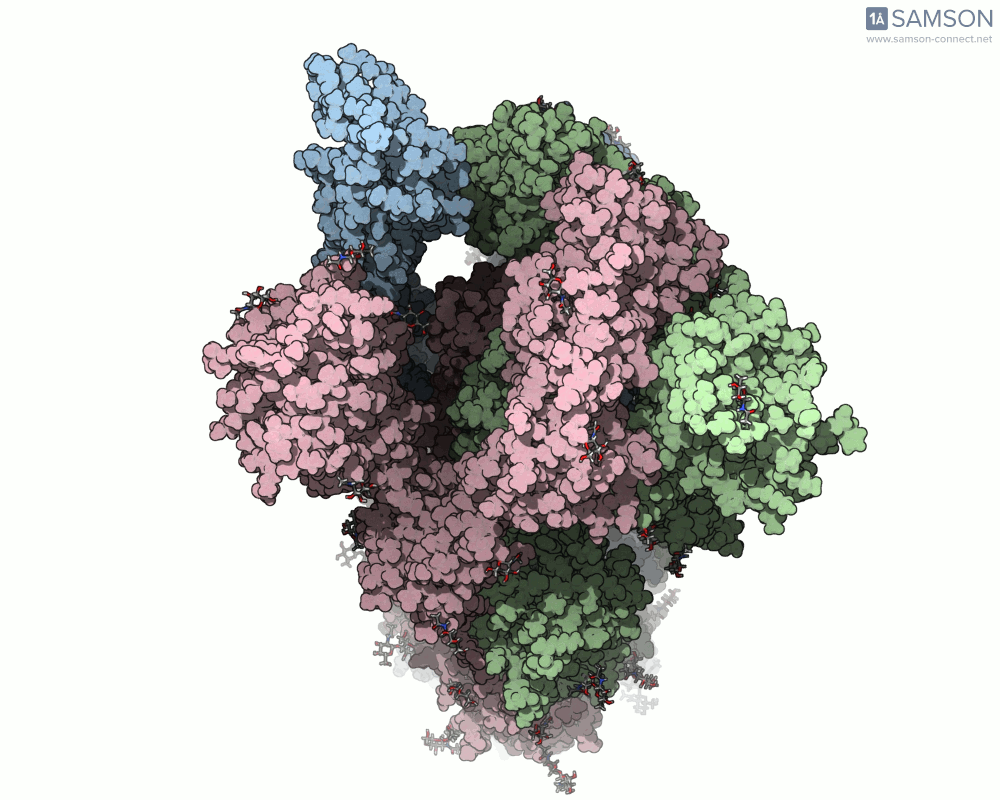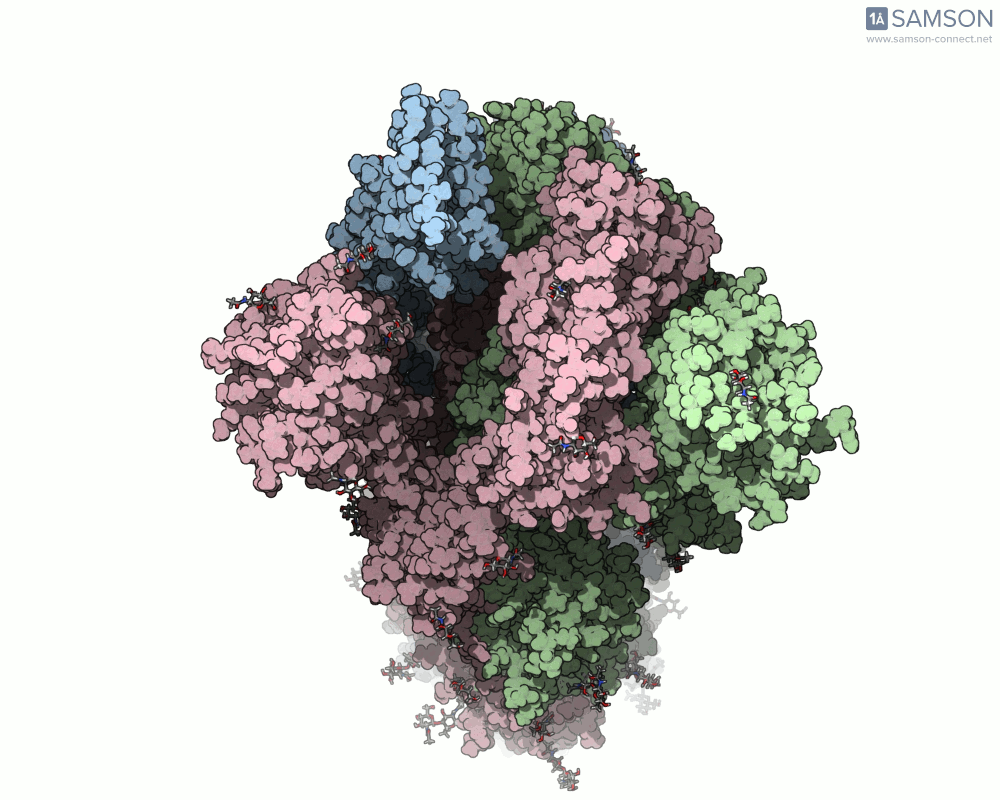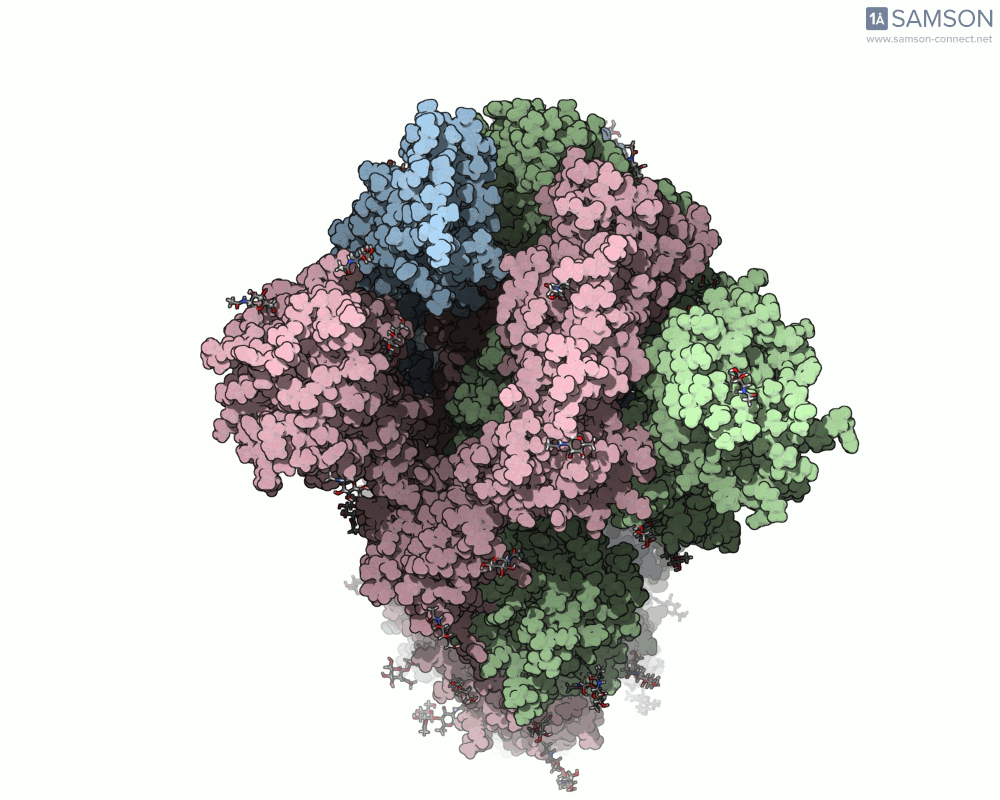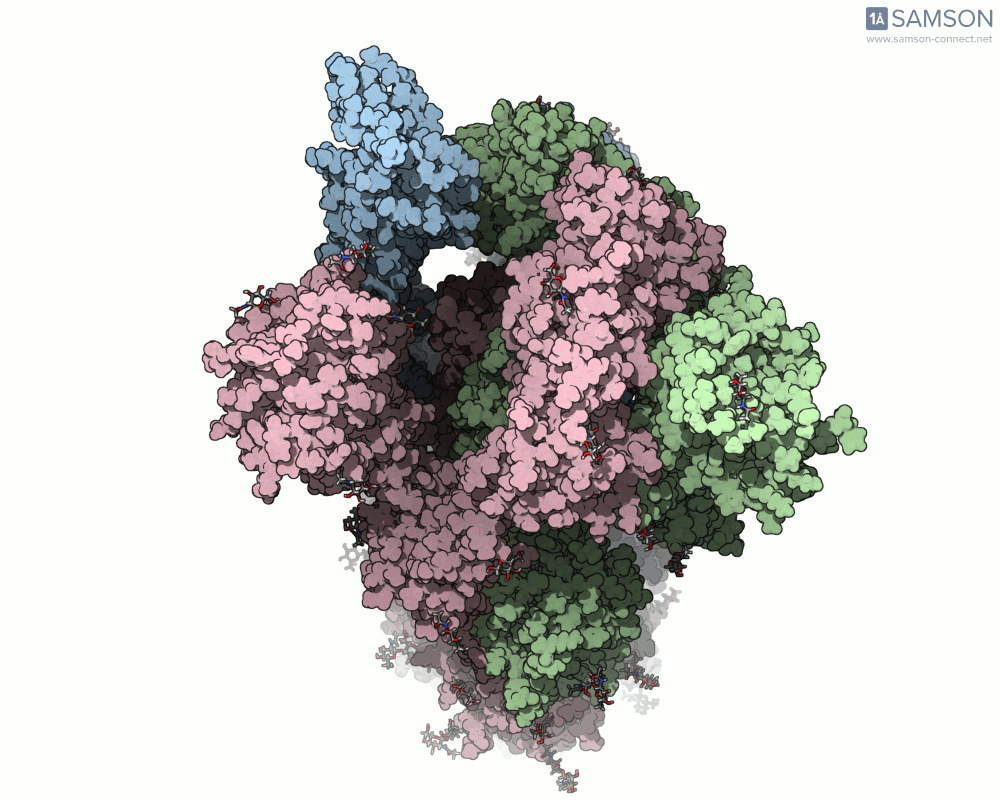
- Top view:
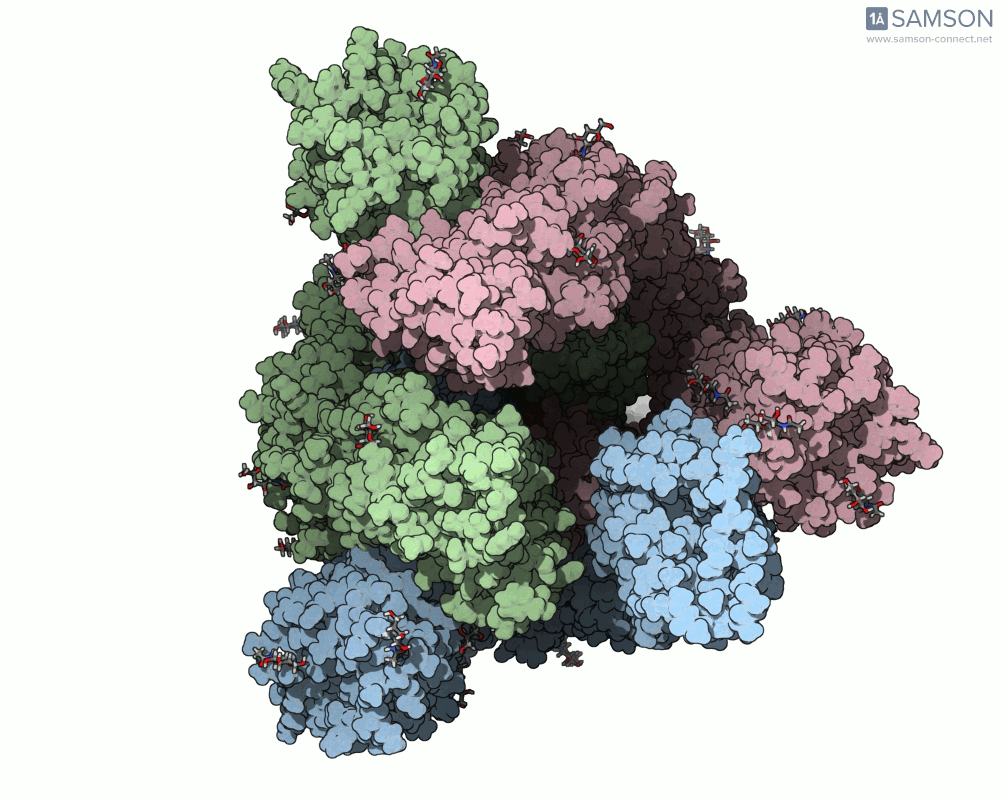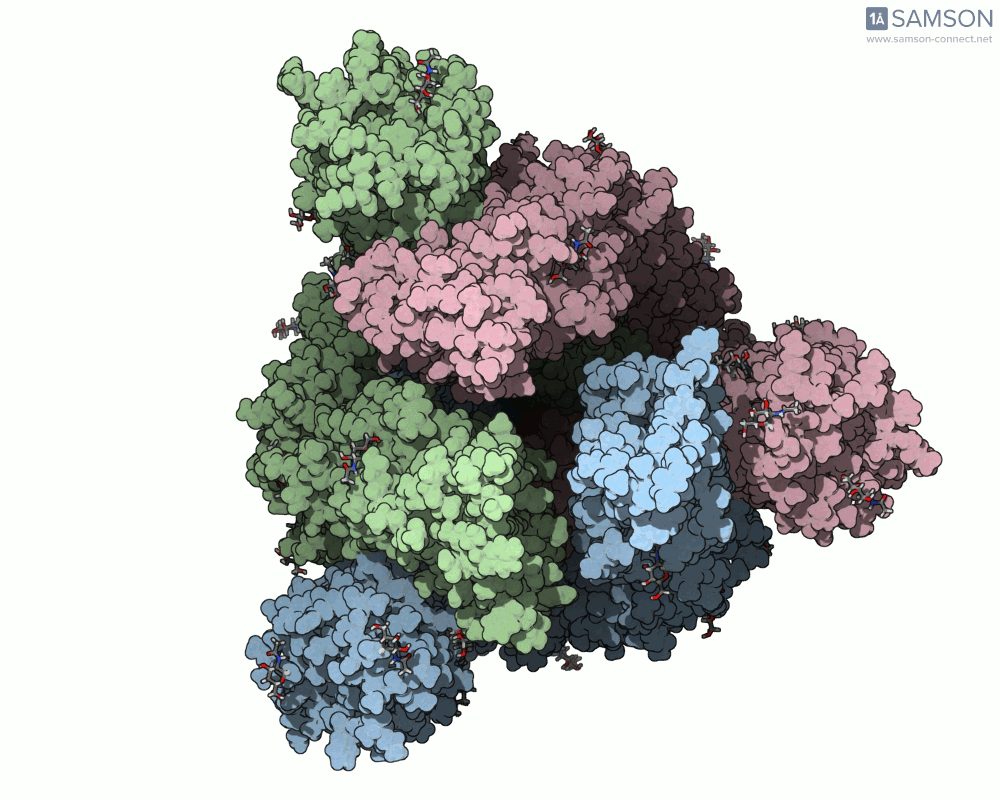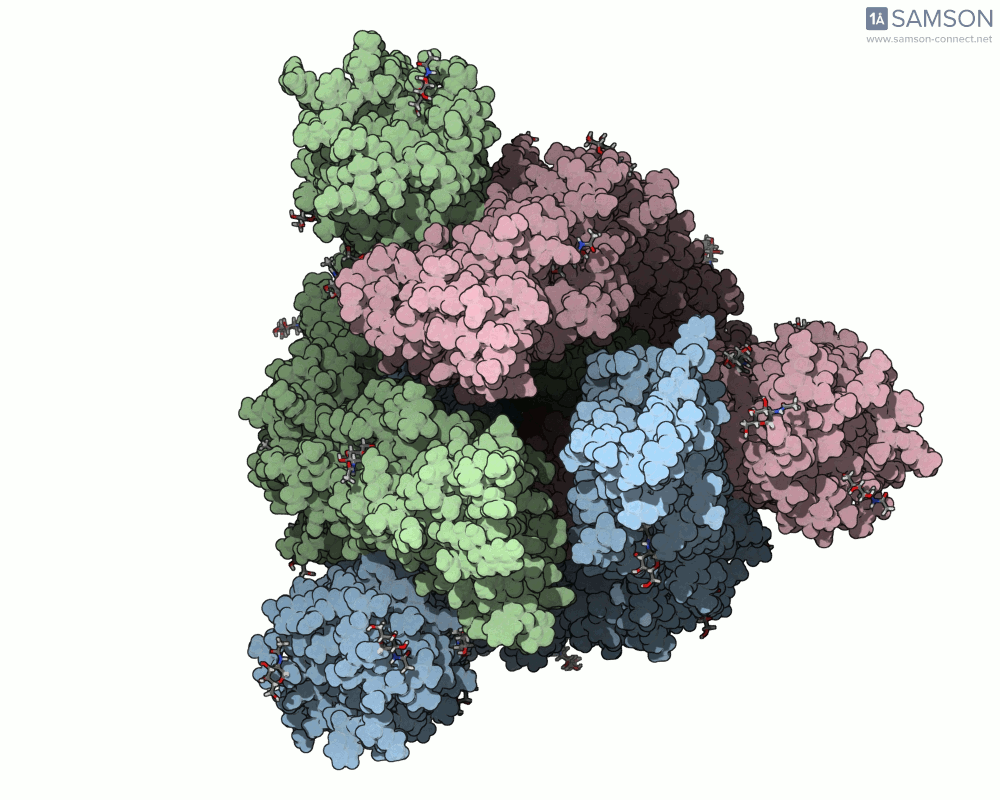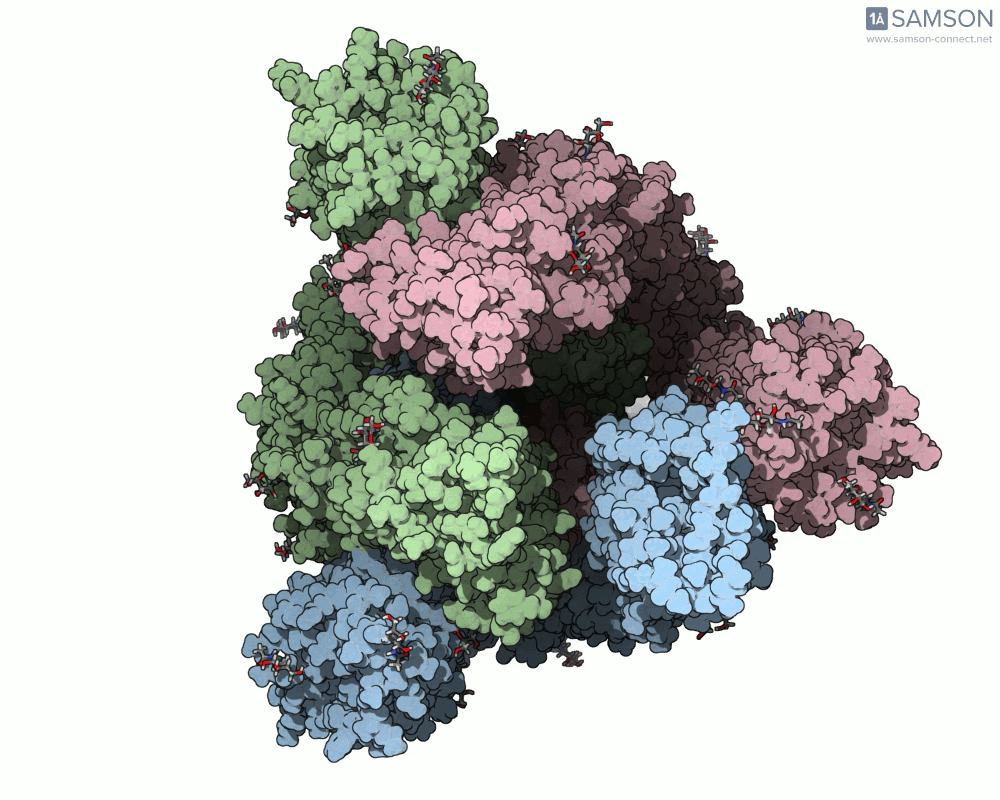
This animation sequence was generated using two SAMSON Extensions: the ARAP Interpolation Path module and the P-NEB (Parallel Nudged Elastic Band) module. The computational path reflects a plausible pathway from closed to open conformations, providing a valuable conceptual framework for modeling and hypothesis testing.
While not experimentally validated, the computed motion can serve as a high-quality starting point for further studies, such as protein-ligand docking, dynamic simulations, or antibody design. Researchers can download the full trajectory in multiple formats, including PDB and SAMSON’s native format, to integrate into their workflows:
Here’s what’s especially useful: the provided .sam file not only gives you the structures and the path but also enables direct animation within SAMSON’s visual interface. Molecular modelers can double-click to start and stop motion, or use specific conformations as boundaries for further simulations.
This is a time-saving resource that molecular modelers can use to:
- Create educational or explanatory diagrams and videos
- Set up more accurate docking scenarios by accounting for structural flexibility
- Explore dynamic epitope exposure for antibody design
- Generate hypotheses on transition-state binding events
Explore the animations and download resources from the original documentation page:
https://documentation.samson-connect.net/tutorials/sars-cov-2/coronavirus-computing-the-opening-motion-of-the-sars-cov-2-spike/
SAMSON and all SAMSON Extensions are free for non-commercial use. To start using SAMSON, visit https://www.samson-connect.net.





