Building realistic lipid environments around membrane proteins can be challenging and time-consuming. Ensuring proper orientation of the molecule, avoiding atom overlaps, adding buffer space… these steps often involve a patchwork of tools and manual adjustments that take hours—even before you begin simulations.
The Molecular Box Builder extension in SAMSON simplifies this process. Let’s look at how you can wrap a protein in a lipid layer in a few systematic steps, directly inside SAMSON. No scripting. No third-party file juggling.
Context: Why Wrap Proteins in Lipid Layers?
Membrane proteins play essential roles in biology and are embedded in lipid bilayers in living cells. Simulating them in such an environment is essential to understand their structure and function. However, creating a realistic lipid layer around a protein has been a tedious task… until now.
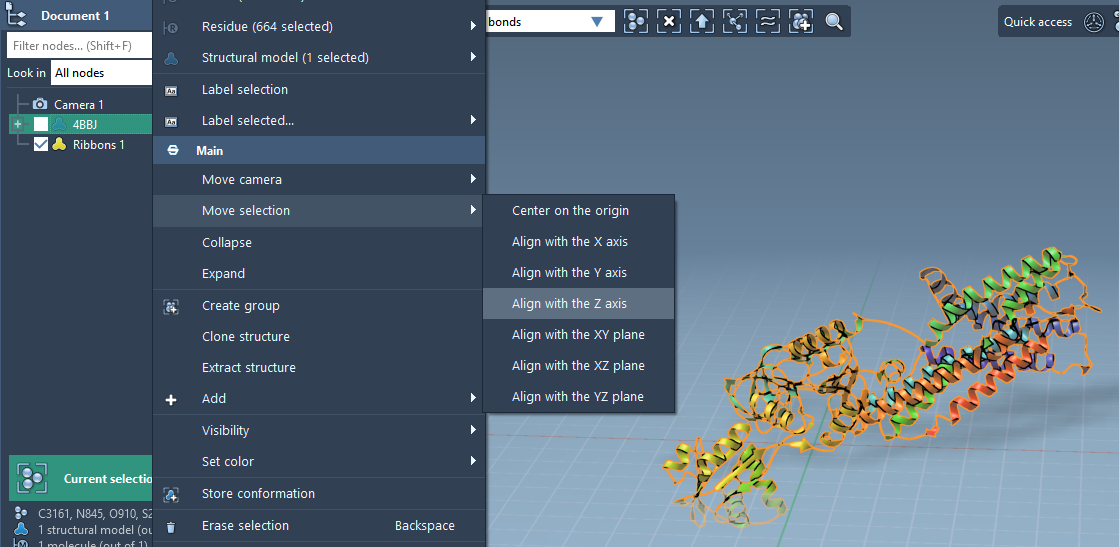
Step 1 – Align the Protein
Start by aligning your protein so the membrane can grow along the correct axis:
- Right-click the protein in the Document view.
- Select Move selection > Align with Z axis.
- Then choose Move selection > Center on the origin.
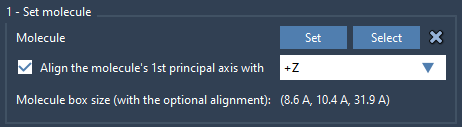
Step 2 – Set the Lipid Molecule
Next, import a lipid (e.g., POPC) that will make up your lipid layer:
- Select the lipid molecule in the scene.
- Open the Molecular Box Builder app and click Set.
- To align the lipid tail with the protein membrane normal, choose alignment to
+Z.
Step 3 – Define the Box
The next step is to define the box where the lipids will be placed:
- Center the box around your aligned protein.
- Adjust the box height to match the thickness of a lipid monolayer and cover enough X-Y area.
- Add a margin if needed to avoid atomic overlaps at generation.
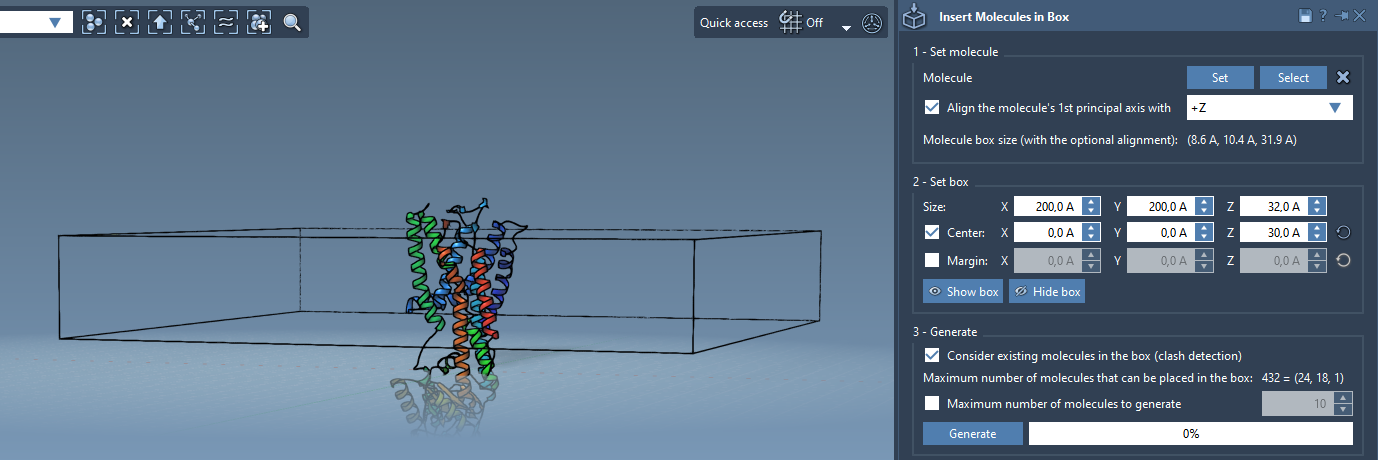
Step 4 – Generate
- Enable Consider existing molecules in the box. This ensures lipids will be inserted only in the available volume, avoiding protein clashes.
- Click Generate. Lipid molecules will be packed around the protein in a few seconds.

Bonus: Building a Bilayer
To build a full lipid bilayer, you can repeat the insertion with inverted alignment:
- First insertion: lipid aligned to
+Z. - Move the box center down along Z.
- Second insertion: use same lipid, but align to
-Z.
The result is a neatly packed, double-sided lipid bilayer around your membrane protein—ready for hydration and simulation.
You can learn more about this workflow in the full Molecular Box Builder documentation.
SAMSON and all SAMSON Extensions are free for non-commercial use. You can get SAMSON at https://www.samson-connect.net.





