If you’ve ever worked with large biomolecular systems, you know the frustration of trying to select a specific type of residues, atoms, or structural groups in a sea of data. Manually clicking or scrolling through complex molecular hierarchies isn’t just inefficient—it introduces the risk of missing or misidentifying important components critical to your simulation or analysis.
Enter the Node Specification Language (NSL) in SAMSON, which significantly simplifies node selection and filtering. One especially helpful feature: using NSL directly in the Document View to filter molecular structures based on precise attributes.
Using NSL to Filter Nodes in the Document View
The Document View in SAMSON lets you visually navigate and manage elements in your molecular document. However, when the document contains thousands—or even millions—of nodes, distinguishing between backbone atoms, ligands, or structural groups becomes time-consuming.
With NSL filtering, you type a query—like n.t sg—into the filter box at the top of the Document View. This command tells SAMSON: “only show structuralGroup-type nodes, please.” Once filtered, you can hit Enter to select them directly.
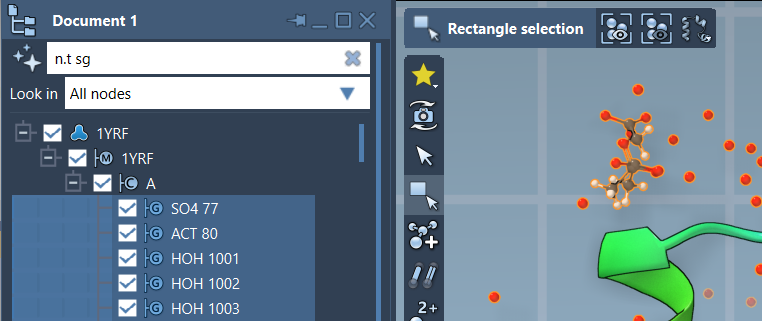
Why This Matters
- Saves time: Skip lengthy clicks and isolate the data you care about in seconds.
- Minimizes errors: Avoid unintentionally including or excluding nodes in your selection.
- Flexible: NSL supports complex queries using logic, topology, and proximity—giving you detailed control.
Common Use Case Examples
Here are a few examples of how to leverage NSL filtering:
n.t r: Show only residue nodesr.id 10:40: Show residues with IDs between 10 and 40n.t sc h S: ShowsideChainnodes that contain at least one sulfur atomr.t ALA: Show only alanine residues
Let the AI Assistant Help
Typing expressions manually can feel daunting, especially if you are new to NSL. Good news: SAMSON includes an AI Assistant that understands your document’s structure. Click the ![]() button next to the filter input field in the Document View. Describe what you want in plain English—like “select all acid residues with more than 5 atoms”—and get an NSL expression instantly.
button next to the filter input field in the Document View. Describe what you want in plain English—like “select all acid residues with more than 5 atoms”—and get an NSL expression instantly.
Summary
Filtering nodes in the Document View with NSL is a small step that makes a big difference in how fluidly you can work on complex molecular models. Whether you’re identifying functional groups, selecting ligands, or isolating nonstandard residues, NSL gives you fast and accurate control. The next time you’re scrolling endlessly through the Document View, try typing n.t r instead—you may never go back.
To explore more NSL features and refine your filter techniques, visit the official NSL documentation page.
SAMSON and all SAMSON Extensions are free for non-commercial use. You can get SAMSON at https://www.samson-connect.net.





