Author: OneAngstrom
Struggling with Flat Molecular Visuals? Ambient Occlusion Might Be the Fix
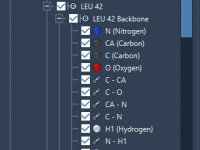
Making Your Molecular Data Speak: Controlling Property Model Visibility in SAMSON
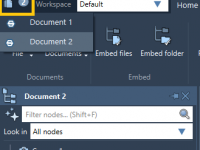
Taming Complex Molecular Structures with SAMSON’s Document View
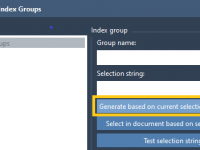
Custom Index Groups in GROMACS Wizard: How and Why to Use Them
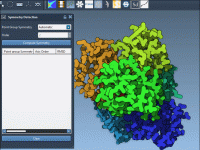
What to Do When Your Protein Has Multiple Symmetries?
From Virtual DNA to Simulation: Exporting Designs from Adenita
Choosing Better Colors for Your Molecular Models with Discrete Palettes in SAMSON
Building Your Own Molecular Modeling Tools in SAMSON Without Starting from Scratch

For many researchers and developers in molecular modeling, one recurring challenge is adapting software tools to specific workflows. Whether you’re modeling new nanostructures, designing drug candidates, or visualizing complex biophysical systems, off-the-shelf features often cover only part of the task.…
Mastering Camera Movements for Molecular Presentations
One of the most overlooked yet essential challenges in molecular modeling presentations is maintaining clarity while guiding viewers through complex 3D structures. Whether you’re creating an educational video, showcasing molecular docking, or simply navigating large biomolecules during a meeting, poor…