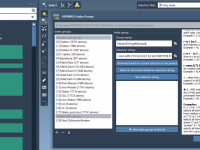
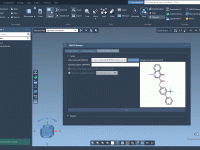
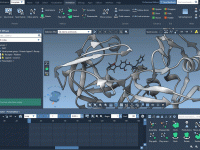
Author: OneAngstrom
How to Keep Your Molecular Labels Clear at Any Zoom Level
From Path to Playback: A Simple Guide to Animating Molecular Trajectories in SAMSON
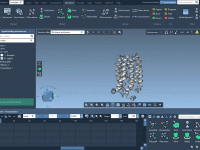
Molecular modelers often generate complex trajectories—whether from structural transitions, docking analyses or dynamics—but presenting these transitions clearly can be difficult. Communicating results, sharing insights with colleagues, or documenting a simulation’s progress often demands more than static images. This is where…