Author: OneAngstrom
A Practical Way to Define Binding Sites for Protein-Ligand Docking in SAMSON
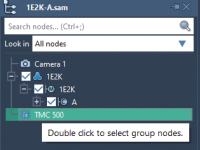
Why Is My Node Missing? Demystifying Visibility in SAMSON
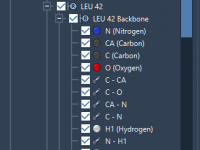
A Quick Way to Find Charged Molecules in Your Molecular Models
When working on complex molecular systems, especially in computational drug design or materials science, one common task is identifying and isolating charged structural models within large datasets. Whether you’re verifying protonation states or selecting compounds for ion-binding studies, efficiently filtering…
Made a Modeling Mistake? Here’s How to Step Back Safely
Managing Complex Molecular Models with the Document View in SAMSON
Populating Lipid Layers Around Proteins in 3D: A Practical Guide
Reserving Your Public Profile and Custom Handle in SAMSON Connect
Tidy Up Your Molecular Projects with Folders in SAMSON
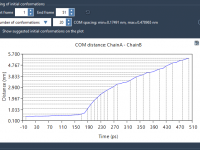
Easily Select Initial Conformations for Umbrella Sampling in SAMSON
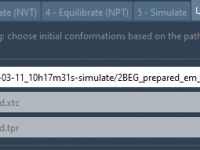
For molecular modelers performing umbrella sampling, generating initial conformations can often feel like a tedious and error-prone process. Whether you’re dealing with long trajectories or a set of predefined conformations, deciding where to extract your snapshots isn’t always straightforward. The…