Author: OneAngstrom
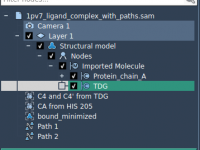
Quickly Target the Right Files in SAMSON Using NSL’s selected Attribute
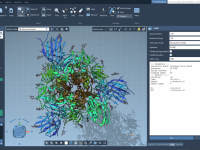
What Do ARAP Edges Actually Represent in Protein Motion?
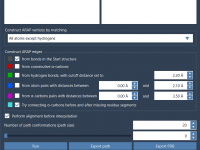
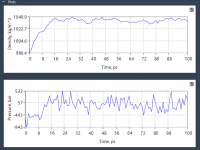
When using ARAP (As-Rigid-As-Possible) Interpolation to generate protein conformational transitions, an essential but sometimes overlooked step is how the structural connectivity—also known as ARAP edges—is defined. For molecular modelers preparing simulations or building transition pathways, getting the connectivity right can…