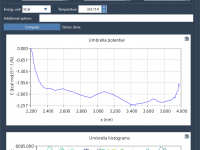
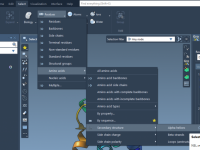
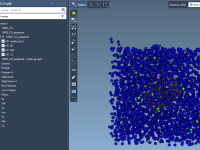
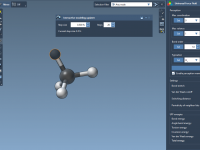
Quickly Explore Molecular Variants with Positional Analogue Scanning in SAMSON
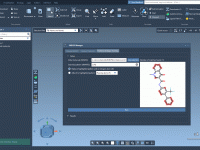
When designing new drug candidates or optimizing a lead compound, molecular modelers often face the challenge of evaluating how small chemical modifications affect binding affinity, selectivity, or physicochemical properties. One effective approach to address this is positional analogue scanning, where…