Author: OneAngstrom
Easier Molecular Storytelling: Set Background Animation in SAMSON
Creating effective molecular presentations is a key part of communicating complex scientific ideas—whether you’re showing binding interactions, illustrating a reaction mechanism, or walking through dynamic conformational changes. However, transitions between animation segments can feel abrupt or visually inconsistent without control…
Editing Protein Conformations Using an Interactive Ramachandran Plot
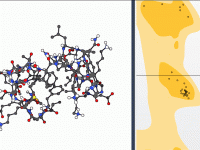
When refining protein structures, one recurring challenge for modelers is ensuring that all residues adopt conformations that are sterically and energetically reasonable. Outliers in dihedral angles can lead to instability in simulations or incorrect biological interpretations. While tools like structure…
Simplifying Molecular Presentation: Docking Atoms with Ease in SAMSON
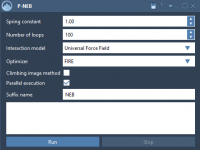
Why Combining Conformations into a Path Saves Time in Transition Path Optimization
Editing Molecular Animations with Track View in SAMSON: A Practical Guide for Researchers
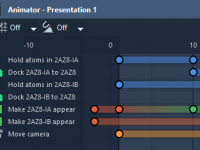
Creating compelling molecular animations can be one of the most time-consuming tasks for researchers preparing publications, presentations, or outreach material. While modern tools offer advanced visualization options, they often lack intuitive ways to manage complex animation timelines, leading to frustration…