Author: OneAngstrom
Cutting Crystals in SAMSON: An Easy Way to Explore Crystal Planes
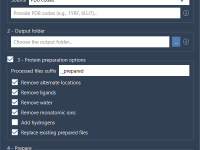
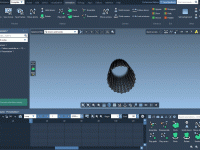
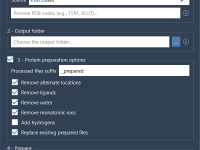
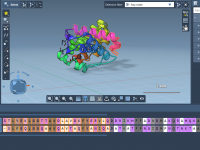
Crystallography plays a vital role in material science, nanotechnology, and structural biology. But even for experienced modelers, visualizing and interacting with crystal structures can feel abstract. A recurring challenge is analyzing how crystals look and behave along specific crystallographic planes…