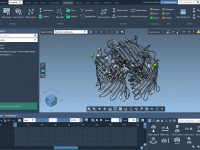
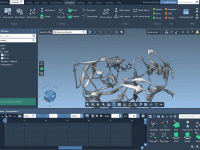
When Less is More: Hiding Atoms to Focus Your Molecular Story
One of the most common challenges in molecular modeling is maintaining clarity in complex scenes. When visualizing large biomolecular systems—like proteins, nucleic acids, or ligands in crowded environments—researchers often find themselves overwhelmed with visual clutter that can obscure important details.…