Author: OneAngstrom
Quickly Find What You Need in Molecular Structures: Using Attribute Filtering in SAMSON
Fast, Consistent Molecular Visuals with Visual Presets in SAMSON
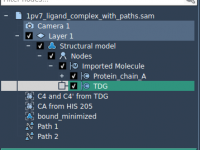
Exporting Ligand Movement Along a Path in SAMSON
Visualizing the SARS-CoV-2 Spike Opening in Motion: Why Animations Matter for Molecular Modelers
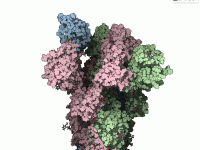
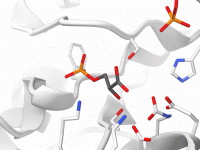
Understanding the conformational changes in viral proteins is crucial when you’re designing inhibitors, vaccines, or simply trying to comprehend viral infectivity mechanisms. One typical challenge for molecular modelers and computational biologists is visualizing these transitions effectively. Static structures in the…
Drawing Carbon Nanotubes with Your Mouse: A Quick Guide
The Frame that Breathes: Adding Pauses in Molecular Presentations
A Smoother Workflow: Predict Protein Structures with AlphaFold-2 in SAMSON
Preventing Setup Headaches: Choosing Inputs and Parameters for GROMACS Simulations in SAMSON
Spend less time tweaking molecular visuals
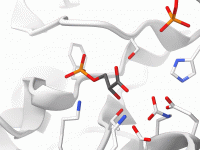
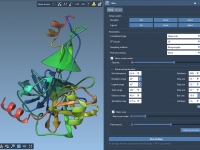
Creating clear, consistent visuals of molecular systems can be a tedious task, whether for scientific communication, teaching, or just keeping track of complex simulations. The common workaround involves repeatedly adjusting representations, color schemes, visibility settings, zoom levels, and more —…