Author: OneAngstrom
Why Your Molecules Look Flat (And How Ambient Occlusion Helps)
Exporting Ligand Trajectories Along Paths: A Practical Guide for Molecular Modelers
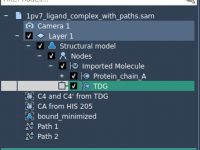
Understanding how ligands move through binding pockets is vital in molecular modeling, whether you’re simulating binding/unbinding events, preparing free energy calculations, or simply visualizing molecular mechanisms. If you’ve ever computed a path (e.g., using a method like parallel nudged elastic…
Quickly Find Conformations by Atom Count in SAMSON
When working with molecular models that have multiple conformations, navigating and managing them efficiently becomes essential—especially when the system’s complexity grows. If you’ve ever wished for a faster way to pinpoint specific conformations within large molecular structures in SAMSON, you’re…
Quickly Find Molecular Conformations by Atom Count in SAMSON
One common challenge in molecular modeling is navigating large datasets of conformations—especially when analyzing or comparing molecular structures with vastly different complexities. Sometimes all a modeler wants is to isolate conformations of similar complexity, or to filter out conformations that…
Make Your Molecular Animations Breathe: How to Pause for Clarity in SAMSON
Building a Lipid Bilayer Around a Protein in Minutes
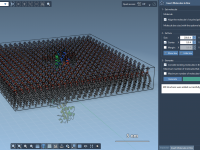
Creating realistic membrane-protein systems is a recurring challenge in molecular modeling. Membrane proteins play critical biological roles, but setting up simulations that combine them with appropriate lipid environments often involves multiple tools, file format conversions, and a fair amount of…