Author: OneAngstrom
Refining Ligand Unbinding Pathways: A Step-by-Step Guide Using P-NEB in SAMSON
Before You Simulate: The Smart Way to Clean Up Molecular Geometries

When preparing molecular systems for simulation, clean geometry isn’t optional—it’s essential. Poor atomic arrangements can lead to unrealistic behavior, convergence failures, or wasted compute time. That’s why optimizing geometries before launching simulations should be part of every molecular modeler’s workflow.…
Keeping Molecules Still for Better Animations
Smoother Transitions in Scientific Presentations: Setting Backgrounds with SAMSON Animator

When preparing molecular presentations, researchers often focus heavily on the scientific content—molecular structures, simulation results, docking poses—and understandably so. However, the presentation layer of that information is also crucial. Transitions that appear abrupt or distracting can reduce clarity or even…
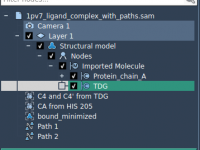
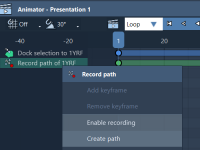
Turning Molecular Animations into Usable Paths
Easier Edits: A Look at DNA Editing Tools in Adenita
Designing DNA nanostructures often involves more than just creating structures — it requires modifying them. Whether you’re untangling a strand, changing connections, or adjusting geometry, manually handling these edits can quickly become error-prone and tedious. Fortunately, Adenita, an extension of…