Author: OneAngstrom
Quickly Toggle Light Node Visibility in Your Molecular Scene
Opening Molecular Pockets Using Normal Modes: A Practical Workflow
Making Molecular Models Fade Away: Using the Disappear Animation in SAMSON
Progressively Hiding Atoms to Highlight Molecular Mechanisms
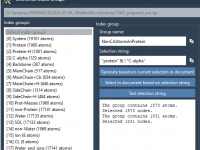
Creating Custom Index Groups in GROMACS Wizard Without Breaking Your Workflow
A Practical Guide to Setting Up Custom Index Groups for Pulling Simulations in GROMACS Wizard
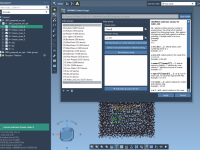
Setting up center-of-mass pulling simulations in GROMACS often requires the definition of custom index groups—particularly when dealing with multi-chain protein systems. For many molecular modelers, this step introduces friction, especially if you’re switching between the command line and visualization tools.…
Position, Click, Animate: A Practical Guide to Keyframe Motion in Molecular Models
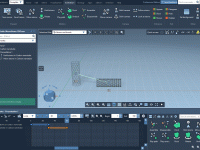
In molecular modeling, dynamic presentations can help clarify complex mechanisms far better than static images. Whether you’re illustrating a potential drug interaction, conformational change, or even just repositioning molecules to highlight geometry, animations go a long way in explaining scientific…