Author: OneAngstrom
Enhancing Molecular Precision with SAMSON’s Simple Script Extension
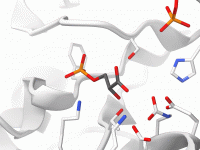
Minimizing NMR Structures Without the Hassle of Custom Force Fields
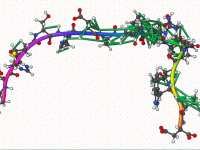
For molecular modelers working with NMR-derived structures, refining these structures into high-quality, publication-ready models without extensive manual setup can be a challenge. Whether you’re preparing protein-ligand complexes or handling non-standard residues, ensuring that your final structures are both chemically valid…
Streamlining Molecular Coordination Exports for Path Analysis
The ability to track and analyze atom trajectories along molecular pathways is an essential tool in molecular modeling. Whether preparing input files for reaction coordinate studies, performing free energy calculations, or studying ligand binding/unbinding pathways, exporting precise atomic coordinates is…
Making Molecular Models Rock with SAMSON
Mastering Segment Attributes for Efficient Molecular Modeling
Understanding Render Preset Attributes in Molecular Visualization
When designing and visualizing molecular models, managing visual representations effectively is key to ensuring accurate communication and understanding of your molecular structures. One helpful feature in SAMSON’s integrative molecular design platform is the ability to work with render preset attributes,…
A Practical Guide to Preparing Coarse-Grained Systems with GROMACS Wizard
Simplifying Molecular Modeling with the SAMSON Interface
Unveiling Light Attributes in Molecular Modeling
For molecular modelers, precise control over scene representation is crucial when designing molecular simulations or visualizations. One common challenge is efficiently tailoring and managing visual elements—such as light nodes—within molecular models. Thankfully, SAMSON’s Node Specification Language (NSL) provides a robust…