Author: OneAngstrom
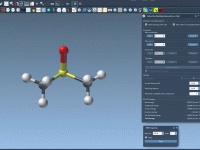
Make Visual Selection Easier in SAMSON with Render Preset Attributes
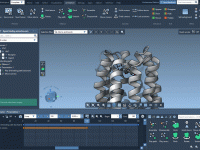
Simulating Multiple Conformations at Once: A Time-Saving Workflow for Molecular Modelers
One of the recurring challenges in molecular modeling is evaluating how different conformations of a single molecular system behave under identical simulation conditions. Whether you’re performing Umbrella Sampling or exploring conformational variability, setting up individual simulations for each structure can…
Keeping Your Focus: Following Atom Groups Without Moving the Camera
Make Molecules Appear Right When You Need Them
When preparing molecular animations for presentations or publications, timing is everything. A common challenge in molecular modeling is making important molecules or structural elements appear exactly when they become relevant—without fading effects or complex scripting. If you’ve ever struggled with…
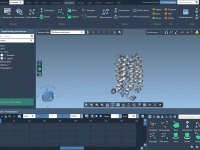
Focus Your Molecular Optimization: How to Minimize a Single Molecule in SAMSON
How to Filter and Organize Your Camera Nodes in SAMSON
When working on complex molecular models or reaction simulations, controlling the viewpoint can be as important as the data itself. Molecular modelers often resort to creating multiple camera nodes to represent different perspectives, highlight specific molecular components, or prepare illustrative…