Avoid Visual Clutter: Disassembling Molecular Structures for Clearer Presentations
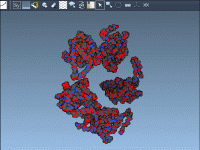
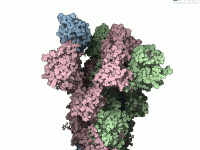
When presenting complex molecular models, especially large assemblies, one common challenge molecular modelers face is visual clutter. Whether you’re showcasing interactions, functional units, or structural domains, it’s easy for your visual narrative to become overwhelming with too many overlapping components.…
Filtering Molecular Backbones by Atomic Composition in SAMSON
A Quick Way to Prepare Ligands for Docking with AutoDock Vina Extended
Revealing Protein Interfaces with Symmetry Mates in SAMSON
Choosing the Best Unit Cell Shape in GROMACS Simulations
A Practical Way to Animate Molecular Transitions Without Complex Simulations
Making Molecular Visualizations Easier with Custom Visual Presets in SAMSON
In molecular modeling, a common challenge is consistently generating clear, informative visualizations—especially when dealing with complex systems such as protein-ligand complexes, solvated structures, or multi-chain systems. Adjusting representations, selecting relevant atoms or molecules, setting color schemes, and choosing visualization models…
Why Pausing at the Right Moment Can Improve Your Molecular Animations
Switching Between Molecular Projects Without Losing Context

For many molecular modelers, juggling multiple molecular systems during a modeling session is the norm, not the exception. Whether you’re comparing ligand binding modes, transferring fragments between structures, or running simulations on several conformations, managing multiple files effectively can be…